
Genetics: From Genes to Genomes
6th Edition
ISBN: 9781259700903
Author: Leland Hartwell Dr., Michael L. Goldberg Professor Dr., Janice Fischer, Leroy Hood Dr.
Publisher: McGraw-Hill Education
expand_more
expand_more
format_list_bulleted
Concept explainers
Textbook Question
Chapter 11, Problem 28P
The figure that follows shows the pedigree of a family in which a completely penetrant, autosomal dominant disease is transmitted through three generations, together with microarray analysis of each individual for a biallelic SNP locus (the alleles are C and T).
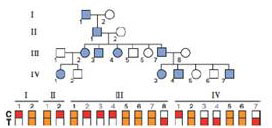
| a. | Do the data suggest the existence of genetic linkage between the SNP locus and the disease locus? If so, what is the estimated genetic distance between the two loci? |
| b. | Calculate the maximum Lod score for linkage between the SNP and the disease locus for this pedigree. What does this value of the Lod score signify? |
Expert Solution & Answer
Trending nowThis is a popular solution!

Students have asked these similar questions
Answer number seven do what it says.
Which of the following is the process that is "capable of destroying all forms of microbial life"?
Question 37 options:
Surgical scrub
Sterilization
Chemical removal
Mechanical removal
After you feel comfortable with your counting method and identifying cells in the various stages of mitosis, use the four images below of whitefish blastula to count the cells in each stage until you reach 100 total cells, recording your data below in Data Table 1. (You may not need to use all four images. Stop counting when you reach 100 total cells.)
After totaling the cells in each stage, calculate the percent of cells in each stage. (Divide total of stage by overall total of 100 and then multiply by 100 to obtain percentage.)
Data Table 1Stage Totals PercentInterphase Mitosis: Prophase Metaphase Anaphase Telophase Cytokinesis Totals 100 100%
To find the length of time whitefish blastula cells spend in each stage, multiply the percent (recorded as a decimal, in other words take the percent number and divide by 100) by 24 hours. (Example: If percent is 20%, then Time in Hours = .2 * 24 = 4.8) Record your data in Data…
Chapter 11 Solutions
Genetics: From Genes to Genomes
Ch. 11 - Choose the phrase from the right column that best...Ch. 11 - Would you characterize the pattern of inheritance...Ch. 11 - Would you be more likely to find single nucleotide...Ch. 11 - A recent estimate of the rate of base...Ch. 11 - If you examine Fig. 11.5 closely, you will note...Ch. 11 - Approximately 50 million SNPs have thus far been...Ch. 11 - Mutations at simple sequence repeat SSR loci occur...Ch. 11 - Humans and gorillas last shared a common ancestor...Ch. 11 - In 2015, an international team of scientists...Ch. 11 - Using PCR, you want to amplify an approximately 1...
Ch. 11 - Prob. 11PCh. 11 - The previous problem raises several interesting...Ch. 11 - You want to make a recombinant DNA in which a PCR...Ch. 11 - You sequence a PCR product amplified from a...Ch. 11 - Prob. 15PCh. 11 - The trinucleotide repeat region of the Huntington...Ch. 11 - Sperm samples were taken from two men just...Ch. 11 - Prob. 18PCh. 11 - a. It is possible to perform DNA fingerprinting...Ch. 11 - On July 17, 1918, Tsar Nicholas II; his wife the...Ch. 11 - The figure that follows shows DNA fingerprint...Ch. 11 - Microarrays were used to determine the genotypes...Ch. 11 - A partial sequence of the wild-type HbA allele is...Ch. 11 - a. In Fig. 11.17b, PCR is performed to amplify...Ch. 11 - The following figure shows a partial microarray...Ch. 11 - Scientists were surprised to discover recently...Ch. 11 - The microarray shown in Problem 25 analyzes...Ch. 11 - The figure that follows shows the pedigree of a...Ch. 11 - One of the difficulties faced by human geneticists...Ch. 11 - Now consider a mating between consanguineous...Ch. 11 - The pedigree shown in Fig. 11.22 was crucial to...Ch. 11 - You have identified a SNP marker that in one large...Ch. 11 - The pedigrees indicated here were obtained with...Ch. 11 - Approximately 3 of the population carries a mutant...Ch. 11 - The drug ivacaftor has recently been developed to...Ch. 11 - In the high-throughput DNA sequencing protocol...Ch. 11 - A researcher sequences the whole exome of a...Ch. 11 - As explained in the text, the cause of many...Ch. 11 - Figure 11.26 portrayed the analysis of Miller...Ch. 11 - A research paper published in the summer of 2012...Ch. 11 - Table 11.2 and Fig. 11.27 together portray the...Ch. 11 - The human RefSeq of the entire first exon of a...Ch. 11 - Mutations in the HPRT1 gene in humans result in at...Ch. 11 - Prob. 44P
Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biology and related others by exploring similar questions and additional content below.Similar questions
- What are Clathrin coated vesicles and what is their function?arrow_forwardHow is a protein destined for the Endoplasmic Reticulum (ER), imported into the ER? Be concise.arrow_forwardFind out about the organisations and the movements aimed at the conservation of our natural resources. Eg Chipko movement and Greenpeace. Make a project report on such an organisation.arrow_forward
- What are biofertilizers and mention the significancearrow_forwardPCBs and River Otters: Otters in Washington State’s Green-Duwamish River have high levels of polychlorinated biphenyls (PCBs) in their livers. PCBs can bind to the estrogen receptors in animals and disrupt the endocrine system of these otters. The PCBs seem to increase the estrogen to androgen ratio, skewing the ratio toward too much estrogen. How would increased estrogen affect the river otter population? Based on your reading of the materials in this unit, what factors can affect fertility in humans? Explain how each of the factors affecting human fertility that you described can disrupt the human endocrine system to affect reproduction.arrow_forwardOther than oil and alcohol, are there other liquids you could compare to water (that are liquid at room temperature)? How is water unique compared to these other liquids? What follow-up experiment would you like to do, and how would you relate it to your life?arrow_forward
- Selection of Traits What adaptations do scavengers have for locating and feeding on prey? What adaptations do predators have for capturing and consuming prey?arrow_forwardCompetition Between Species What natural processes limit populations from growing too large? What are some resources organisms can compete over in their natural habitat?arrow_forwardSpecies Interactions Explain how predators, prey and scavengers interact. Explain whether predators and scavengers are necessary or beneficial for an ecosystem.arrow_forward
- magine that you are conducting research on fruit type and seed dispersal. You submitted a paper to a peer-reviewed journal that addresses the factors that impact fruit type and seed dispersal mechanisms in plants of Central America. The editor of the journal communicates that your paper may be published if you make ‘minor revisions’ to the document. Describe two characteristics that you would expect in seeds that are dispersed by the wind. Contrast this with what you would expect for seeds that are gathered, buried or eaten by animals, and explain why they are different. (Editor’s note: Providing this information in your discussion will help readers to consider the significance of the research).arrow_forwardWhat is the difference between Uniporters, Symporters and Antiporters? Which of these are examples of active transport?arrow_forwardWhat are coupled transporters?arrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage LearningCase Studies In Health Information ManagementBiologyISBN:9781337676908Author:SCHNERINGPublisher:Cengage
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage LearningCase Studies In Health Information ManagementBiologyISBN:9781337676908Author:SCHNERINGPublisher:Cengage Human Biology (MindTap Course List)BiologyISBN:9781305112100Author:Cecie Starr, Beverly McMillanPublisher:Cengage Learning
Human Biology (MindTap Course List)BiologyISBN:9781305112100Author:Cecie Starr, Beverly McMillanPublisher:Cengage Learning Principles Of Radiographic Imaging: An Art And A ...Health & NutritionISBN:9781337711067Author:Richard R. Carlton, Arlene M. Adler, Vesna BalacPublisher:Cengage Learning
Principles Of Radiographic Imaging: An Art And A ...Health & NutritionISBN:9781337711067Author:Richard R. Carlton, Arlene M. Adler, Vesna BalacPublisher:Cengage Learning Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College
Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College

Human Heredity: Principles and Issues (MindTap Co...
Biology
ISBN:9781305251052
Author:Michael Cummings
Publisher:Cengage Learning

Case Studies In Health Information Management
Biology
ISBN:9781337676908
Author:SCHNERING
Publisher:Cengage

Human Biology (MindTap Course List)
Biology
ISBN:9781305112100
Author:Cecie Starr, Beverly McMillan
Publisher:Cengage Learning


Principles Of Radiographic Imaging: An Art And A ...
Health & Nutrition
ISBN:9781337711067
Author:Richard R. Carlton, Arlene M. Adler, Vesna Balac
Publisher:Cengage Learning

Concepts of Biology
Biology
ISBN:9781938168116
Author:Samantha Fowler, Rebecca Roush, James Wise
Publisher:OpenStax College
Mitochondrial mutations; Author: Useful Genetics;https://www.youtube.com/watch?v=GvgXe-3RJeU;License: CC-BY