Becker's World of the Cell (9th Edition)
9th Edition
ISBN: 9780321934925
Author: Jeff Hardin, Gregory Paul Bertoni
Publisher: PEARSON
expand_more
expand_more
format_list_bulleted
Concept explainers
Textbook Question
Chapter 18, Problem 18.5PS
Locating Promoters. The following table provides data concerning the effects of various deletions in a eukaryotic gene encoding 5S rRNA on the ability of this gene to be transcribed by RNA polymerase III.
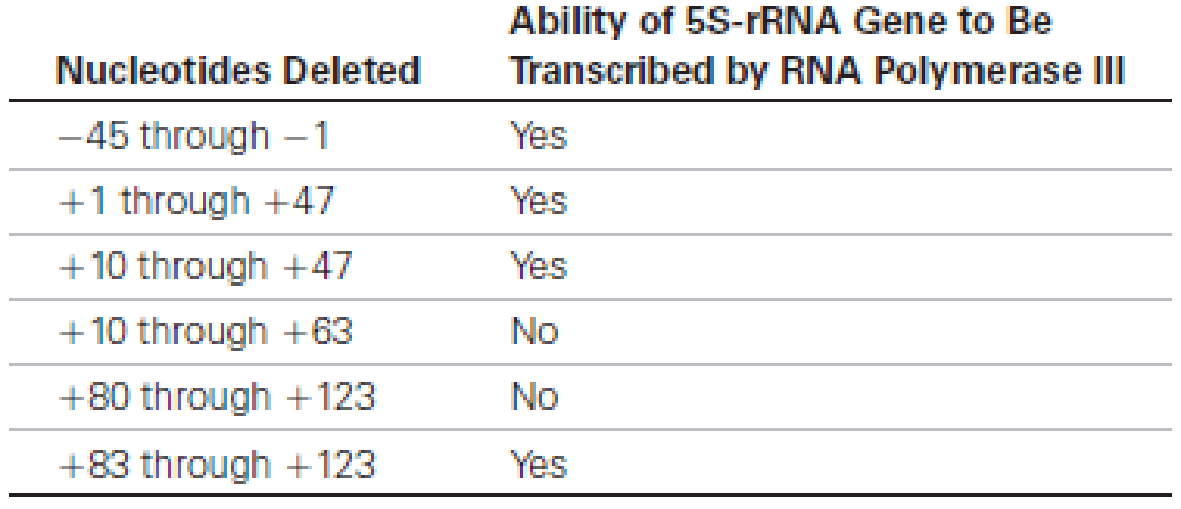
(a) What do these data tell you about the probable location of the promoter for this particular 5S-rRNA gene?
(b) If a similar experiment were carried out for a gene transcribed by RNA polymerase I, what kinds of results would you expect?
(c) If a similar experiment were carried out for a gene transcribed by RNA polymerase II, what kinds of results would you expect?
Expert Solution & Answer
Want to see the full answer?
Check out a sample textbook solution
Students have asked these similar questions
E32. In the technique of DNase I footprinting, the binding of a protein
to a region of DNA protects that region from digestion by DNase I
by blocking the ability of DNase I to gain access to the DNA. In
the DNase I footprinting experiment shown here, a researcher
began with a sample of cloned DNA 400 bp in length. This DNA
contained a eukaryotic promoter for RNA polymerase II. The
assembly of general transcription factors and RNA polymerase II
at the core promoter is described in Chapter 12 (see Figure 12.14).
For the sample loaded in lane 1, no proteins were added. For the
sample loaded in lane 2, the 400-bp fragment was mixed with
RNA polymerase II plus TFIID and TFIIB.
2
400
350
250
175
50
Which region of this 400-bp fragment of DNA is bound by RNA
polymerase II and TFIID and TFIIB?
||
III ||| | ||||
Part I. Structure-Function Relationships in Genes
1. Consider the "two-line model" of a gene shown below - each line represents one
strand of a DNA double helix, and the transcription start site is indicated as +1. Use the
two-line models provided when answering the following questions.
3'
5'
+1
Assume that you know RNA polymerase will move to the right during transcription.
On the diagram above, do the following:
• Label "upstream" and "downstream" on this gene
• Label where you would find the promoter
min
I
• Draw a box where you would expect to find the TATA box
• Draw a third line below the model representing the RNA transcript (label the
ends!)
• Label one of the DNA strands as the template strand
3'
2. Now, let's try that again! This time assume that you know RNA polymerase will move
to the left during transcription. Repeat the same tasks as before on the diagram below:
5'
5'
3'
+1
I
I
5'
3'
E22. The method of Northern blotting is used to determine the amount
and size of a particular RNA transcribed in a given cell type.
Alternative splicing (discussed in Chapter 12) produces mRNAs
of different lengths from the same gene. The Northern blot shown
here was made using a DNA probe that is complementary to the
MRNA encoded by a particular gene. The mRNA in lanes 1 through
4 was isolated from different cell types, and equal amounts of total
cellular MRNA were added to each lane.
2
3
4
Lane 1: MRNA isolated
from nerve cells
Lane 2: MRNA isolated
from kidney cells
Lane 3: MRNA isolated
from spleen cells
Lane 4: MRNA isolated
from muscle cells
Explain these results.
| |
Chapter 18 Solutions
Becker's World of the Cell (9th Edition)
Ch. 18 - Suppose a triplet on the template strand of a...Ch. 18 - Of these three techniques, which one provides the...Ch. 18 - Compare and contrast bacterial and eukaryotic...Ch. 18 - The autoimmune disease systemic lupus...Ch. 18 - QUANTITATIVE Triplets or Sextuplets? In his Nobel...Ch. 18 - The Genetic Code in a T-Even Phage. A portion of a...Ch. 18 - Frameshift Mutations. Each of the mutants listed...Ch. 18 - Prob. 18.4PSCh. 18 - Locating Promoters. The following table provides...Ch. 18 - Prob. 18.6PS
Ch. 18 - Starting Up. Refer to Figure 18-30, which depicts...Ch. 18 - RNA Processing. The three major classes of RNA...Ch. 18 - Prob. 18.9PSCh. 18 - Antibiotic Inhibitors of Transcription. Rifamycin...Ch. 18 - Prob. 18.11PSCh. 18 - Cloning Conundrum. Using established recombinant...Ch. 18 - Nucleoli. Indicate whether each of the following...
Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biology and related others by exploring similar questions and additional content below.Similar questions
- Yes or no only. rna seq can provide sequence and expression data do riboprobes synthesize bu in vitro transcription? does rna causes mutations and lose of function of specific genes?arrow_forwardTranscription AttenuationHow would transcriptionof the E. coli trp operon be affected by the following manipulations of the leader region of the trp mRNA?(a) Increasing the distance (number of bases) betweenthe leader peptide gene and sequence 2(b) Increasing the distance between sequences 2 and 3(c) Removing sequence 4(d) Changing the two Trp codons in the leader peptidegene to His codons(e) Eliminating the ribosome-binding site for the genethat encodes the leader peptide(f) Changing several nucleotides in sequence 3 so thatit can base-pair with sequence 4 but not with sequence 2arrow_forwardI am more confused. how about we start from begining, you post answers on here, and then we go from there? 1. Identify the open reading frame in the following DNA sequence, the protein that this gene encodes for, its function, and the source. 2. "Look carefully at the DNA sequence and identify the start site for transcription" 3. Click on the DNA sequence from the start site of transcription, select all of the sequence, and copy the sequence. Go to the National Center for Biotechnology Information (NCBI) website http://www.ncbi.nlm.nih.gov/. Click on BLAST on the right-hand side under “Popular Resources.” BLAST is a program that will allow you to find the protein sequence for the DNA sequence (gene) you submit. Next click on blastx (translated nucleotide protein). Paste the DNA sequence into the box under “Entry Query Sequence.” Scroll down and click BLAST. The search may take a few seconds; the page will keep updating until the search is completed. You do not need to enter any…arrow_forward
- Plz answer ASAP. I will thumb up You are studying the regulation of the lactose operon in Escherichia coli, by measuring expression of the lacZ gene (i.e production of beta-galactosidase).(a) You identify several loss-of-function mutations in which lacZ is never expressed, in the presence and absence of glucose and lactose. What components of the lac operon could be mutated to produce this phenotype? List all possibilities.arrow_forwardRNA transcription reach low error rate under non-equilibrium steady state, what is the energy source to drive transcription ?arrow_forwardList and briefly explain. C-terminal domain of RNA polymerase II function to ensure that the varoius sets of mRNA processing enzymes carry out their duties at the apporpiate time and place?arrow_forward
- . You have identified a mutation in yeast, a unicellular eukaryote, that prevents the capping of the 5′ end of theRNA transcript. However, much to your surprise, all theenzymes required for capping are normal. You determine that the mutation is, instead, in one of the subunitsof RNA polymerase II. Which subunit is mutant, andhow does this mutation result in failure to add a cap toyeast RNA?arrow_forward. Why is a nonsense suppressor tRNATyr, even though ithas a mutant anticodon that cannot recognize a tyrosinecodon, charged with tyrosine by Tyr tRNA synthetase?arrow_forwardPlease do answer all the questions. I'll definitely give a like You discovered a halophilic bacterium and want to characterize the mechanism involved in producing mature tRNA molecules from larger tRNA precursors. You isolated a large complex composed of a protein component and an RNA component that is capable of cleaving the larger tRNA precursor. To determine which one of the two components is responsible for catalysis, you perform an in vitro tRNA cleavage assay in the proper buffer conditions, including a low concentration of Mg2+ and 0.5 M bovine serum albumin (BSA). BSA is not specific for this reaction. The table below summarizes the results after performing eight separate reactions. The + symbol indicates the included reaction components. Q. Based on the results obtained, what can you conclude about the composition of the biological catalyst required for the maturation of tRNA? Q. Indicate which reactions helped you make your conclusion. Why? Q. Which reactions allowed you…arrow_forward
- . Eukaryotic processing of hnRNA into mature mRNA includes all of the following steps except: ribosome attachment of methionine to the 5’-AUG-3’ codon 5’-addition of a 7-methylguanosine cap 3’-addition of a polyadenylated tail splicing together of exons excision of intronsarrow_forward. Recall that the trp operon has a special leader sequence (trpL) between the operator and the structural genes that offers attenuation as a mechanism for regulation of gene expression. (A) Draw a diagram of a trpL region of the operon when tryptophan is abundant in the cell.Label the following features: the DNA, 5’ and 3’ polarity of the RNA, the regions 1, 2, 3,and 4 and poly-U of the RNA, the pair of Trp codons (UGG), the ribosome, and RNA-Pol,along with any stem-loop structure that would form under these conditions (B) In the above example, will the rest of the trp operon genes be expressed? Briefly describe your reasoning why or why not (C) The trp codons in region 1 of the trpL gene have mutated to cysteines (UGG to UGC). What will be the effect on attenuation gene regulation of the trp operon? Brieflyexplain your reasoning.arrow_forwardGACA CAAT box A site P site E site DREASAARE III III III III III factors and RNA pol to bind to in order to activate gene expression. Accepts charged tRNA molecules whose anticodon complements the codons of mRNA. Holds a nascent elongating polypeptide before final release. Unique sequence found in bacteria between TATAAT and START. 5 upstream consensus motif that is easily melted for transcription purposes in prokaryotes.arrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning

Biochemistry
Biochemistry
ISBN:9781305577206
Author:Reginald H. Garrett, Charles M. Grisham
Publisher:Cengage Learning
QCE Biology: Introduction to Gene Expression; Author: Atomi;https://www.youtube.com/watch?v=a7hydUtCIJk;License: Standard YouTube License, CC-BY