
Study Guide for Campbell Biology
11th Edition
ISBN: 9780134443775
Author: Lisa A. Urry, Michael L. Cain, Steven A. Wasserman, Peter V. Minorsky, Jane B. Reece, Martha R. Taylor, Michael A. Pollock
Publisher: PEARSON
expand_more
expand_more
format_list_bulleted
Concept explainers
Textbook Question
Chapter 16, Problem 5IQ
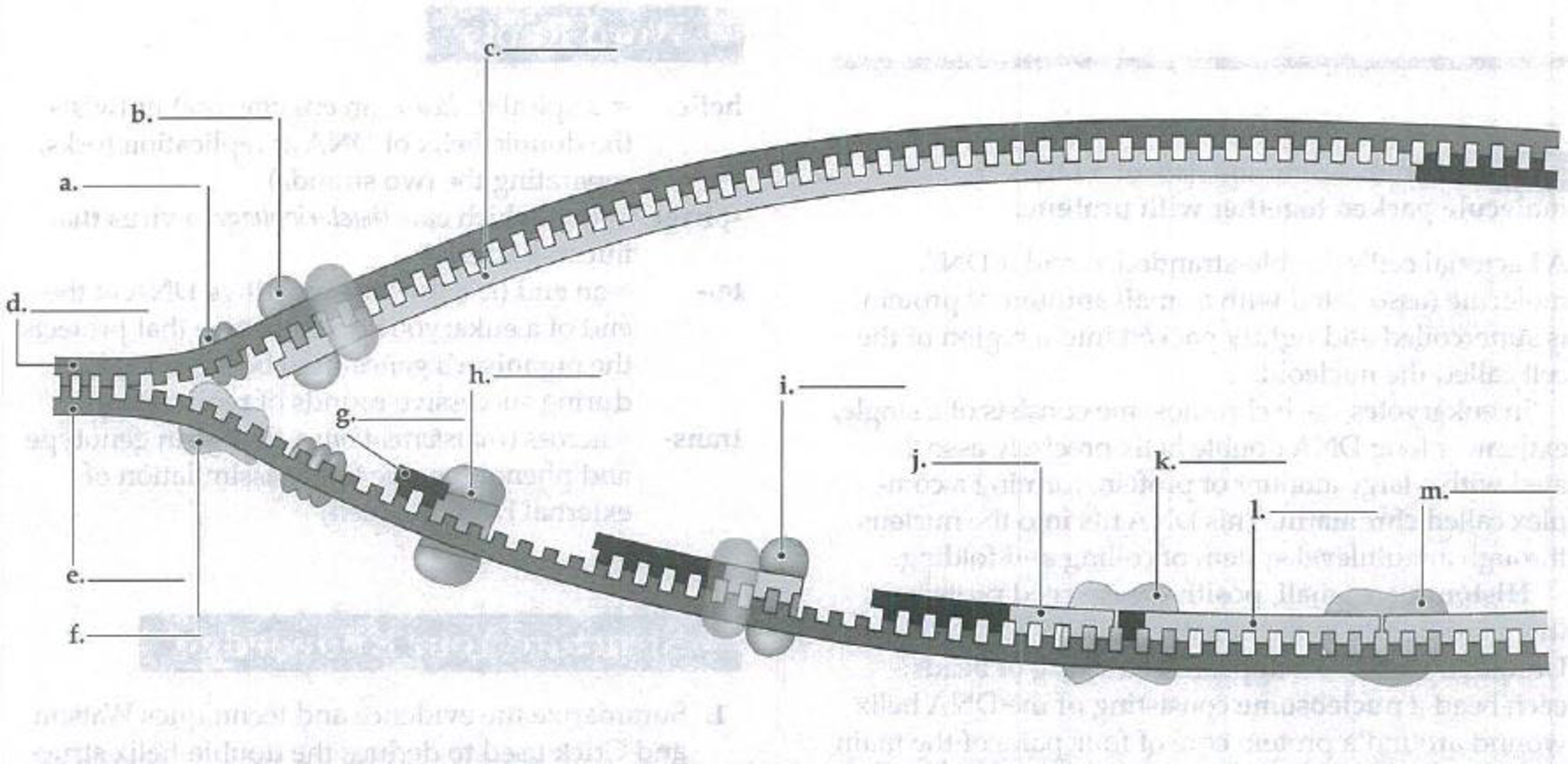
In this diagram of bacterial
Expert Solution & Answer
Want to see the full answer?
Check out a sample textbook solution
Students have asked these similar questions
Define the following words: replication fork, leading strand, lagging strand, Okazaki fragments, helicase, single strand, binding protein, primase, DNA polymerase 3, DNA polymerase 1, dna ligase and nuclease
Given the diagram of the replication fork below,
indicate the chemical group (5'-P, 3'-P, 3'-OH or 5'-OH)
most likely to be found at the sites indicated below by the dots labeled A, B, and C.
Sort the phrases into the appropriate bins depending on which protein they describe.
1) Binds at the replication fork
2) binds after the replication fork
3) binds ahead of the replication fork
4) breaks H-bonds between bases
5) prevents H-bonds between bases
6) breaks covalent bonds in DNA backbone
Helicase:
Topoisomerase:
Single-strand binding protein:
Chapter 16 Solutions
Study Guide for Campbell Biology
Ch. 16 - Hershey and Chase devised an experiment using...Ch. 16 - Review the structure of DNA by labeling the...Ch. 16 - Using different colors for heavy (parental) and...Ch. 16 - Look back to Interactive Question 16.2 and label...Ch. 16 - In this diagram of bacterial DNA replication,...Ch. 16 - Draw the last Okazaki fragment being formed on the...Ch. 16 - List the successive levels of packing in a...Ch. 16 - Prob. 1SYKCh. 16 - Prob. 2SYKCh. 16 - One of the reasons most scientists thought...
Ch. 16 - Transformation involves a. the uptake of external...Ch. 16 - Prob. 3TYKCh. 16 - Which of the following most closely represents...Ch. 16 - Prob. 5TYKCh. 16 - Prob. 6TYKCh. 16 - In their classic experiment, Meselson and Stahl a....Ch. 16 - The joining of nucleotides in the polymerization...Ch. 16 - DNA polymerase is not able to begin copying a DNA...Ch. 16 - Prob. 10TYKCh. 16 - Prob. 11TYKCh. 16 - Prob. 12TYKCh. 16 - Which of the following is least related to the...Ch. 16 - Prob. 14TYKCh. 16 - Prob. 15TYKCh. 16 - Prob. 16TYKCh. 16 - Prob. 17TYKCh. 16 - Which of the following statements about telomeres...Ch. 16 - You are trying to test your hypothesis that DNA...Ch. 16 - Given the experimental procedure explained in...Ch. 16 - Prob. 21TYKCh. 16 - Biologists have learned from the technique of...
Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biology and related others by exploring similar questions and additional content below.Similar questions
- Define the following terms related to DNA replication: origin of replication, helicase, single-strand binding proteins, topoisomerase, primase, RNA primer, DNA polymerase III, DNA polymerase I, and DNA ligase.arrow_forwardThe proteins and enzymes listed below are all required for DNA replication in E. coli, but they are listed in a random order. Determine the correct order in which they function in replication, by selecting the correct number from the drop-down menu in each case, with 1 being first and 6 being last.arrow_forwardA region of DNA has six copies of a trinucleotide repeat. During one round of replication, the template strand slips as shown in the diagram. How many repeats will the DNA have if the newly synthesized strand is used as a template in the next round of replication? 1 5'-CAG 3'-GTC GTC 4 2 GTCH 3 2 CAG CAG GTC 3 GTC 5 4 CAG-3' GTC -5' 6 Next round of DNA replicationarrow_forward
- DNA is made of two strands that are antiparallel. If one strand runs from 3’ to 5’ direction the other one will go from 5’ to 3’ direction. During replication or transcription, whatever the process is, it will always follow the 5’ to 3’ direction using the 3’ to 5’ directed strand as the template strand. Therefore, if following is the DNA sequence5’-CCG ATC GCA CAA-3’a) Using this sequence as template after transcription no protein can be translated. Why? I. Presence of start codonII. Absence of start codonIII. Due to mutationb) If you want to start the translation, what change you need in the second codon (from 5’ to 3’ direction)?I. Substitution of C with GII. No changeIII. Deletion of CIV. Both I & IIIarrow_forwardProvide the complementary strand and the RNA transcription product for the following DNA template segment:5'-AGGGGCCGTTATCGTT-3'arrow_forwardDiscuss DNA replication of prokaryotes and please mention all of the enzymes and components listed below. DNA Primase – DNA directed “RNA Pol” which inserts nucleoside triphosphates (NTPs) Primers are oligonucleotides; priming process is the formation of primers DNA Helicase – separates the DNA in advance of the replication fork (in E. coli DNA Helicase II); binds at AT-rich region of DNA; ATP then binds the helicase Single-stranded DNA-binding Protein (SSBP) – no enzymatic activity; does not consume ATP Topoisomerase – alter the supercoiling of double-stranded DNA DNA Ligase – nicking of strands done for replication to continue Okazaki fragments DNA Polymerase – removes primer via 5’ to 3’ exonuclease activity; comes again for 5’ to 3’ polymerization activity (closes the gap between Okazaki fragments) Prokaryotes DNA Pol I – auxiliary enzyme to DNA Pol III; repairs damage; capable of excising pyrimidine dimers; polymerization via single active site that can bind all 4 dNTPs;…arrow_forward
- DNA polymerase occasionally incorporates the wrong nucleotide during DNA replication. If left unrepaired, the base-pair mismatch that results will lead to mutation in the next replication. As part of a template strand, the incorporated wrong base will direct the incorporation of a base complementary to itself, so the bases on both strands of the DNA at that position will now be different from what they were before the mismatch event. The MER-minus strain of yeast does not have a functional mismatch excision repair system, but it has normal base excision repair and nucleotide excision repair systems. Which of the following statements is correct about differences in the mutation spectrum between MER-minus and wildtype yeast? More than one answer is correct. Options: More point mutations will arise in MER-minus yeast. Fewer point mutations will arise in MER-minus yeast as compared with wildtype. Of the total point mutations that…arrow_forwardDNA polymerase occasionally incorporates the wrong nucleotide during DNA replication. If left unrepaired, the base-pair mismatch that results will lead to mutation in the next replication. As part of a template strand, the incorporated wrong base will direct the incorporation of a base complementary to itself, so the bases on both strands of the DNA at that position will now be different from what they were before the mismatch event. The MM-minus strain of yeast does not have a functional mismatch excision repair system, but it has normal base excision repair and nucleotide excision repair systems. Which of the following statements is correct about differences in the mutation spectrum between MMR-minus and wildtype yeast? More than one answer is correct. (a) More point mutations will arise in MMR-minus yeast. Fewer point mutations will arise in MMR-minus yeast as compared with wildtype. (b) Of the total point mutations that do occur, the fraction in which G is replaced by C will be…arrow_forwardDNA polymerases are processive, which means that they remain tightly associated with the template strand while moving rapidly and adding nucleotides to the growing daughter stand. Which piece of the replication machinery accounts for this characteristic? Helicase Sliding Clamp Single Stranded Binding Protein Primasearrow_forward
- The figure below depicts various elements of the eukaryotic replication machinery in action. Enter the name for the protein depicted by each box. Box A Box B Box C Box D Box E Box F DNA polymerase on lagging strand (just finishing an Okazaki fragment) F Maintains polymerase association with DNA Enzyme extends separation of DNA strands Synthesizes RNA fragments that hybridize to DNA Relaxes supercoiled DNA ahead of replication fork Maintains DNA is single stranded state Promotes binding of processivity factors to DNA Newly synthesized strand pocoar Leading-strand template A New Okazaki fragment RNA primer E Lagging-strand template DNA polymerase on leading strand B C D Saaragon - Next Okazaki fragment will start here Parental DNA helixarrow_forwardDNA is made of two strands that are antiparallel. If one strand runs from 3’ to 5’ direction the other one will go from 5’ to 3’ direction. During replication or transcription, whatever the process is, it will always follow the 5’ to 3’ direction using the 3’ to 5’ directed strand as the template strand. Therefore, if following is the DNA sequence 5’-CCG ATC GCA CAA-3’ Using this sequence as template after transcription no protein can be translated. Why? Presence of start codon Absence of start codon Due to mutation If you want to start the translation, what change you need in the second codon (from 5’ to 3’ direction)? Substitution of C with G No change4 Deletion of Both I & IIIarrow_forwardWhat is unique about the nature of the replication of the bacterial chromosome? Chromosome is bidirectional and has a fixed point of initiation Chromosome is unidirectional or reciprocal Chromosome is unidirectional and has a fixed point of initiation Chromosome is multirepliconic but not telomeric Chromosome is bidirectional or multirepliconicarrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning

Human Heredity: Principles and Issues (MindTap Co...
Biology
ISBN:9781305251052
Author:Michael Cummings
Publisher:Cengage Learning
QCE Biology: Introduction to Gene Expression; Author: Atomi;https://www.youtube.com/watch?v=a7hydUtCIJk;License: Standard YouTube License, CC-BY