
Human Anatomy & Physiology (11th Edition)
11th Edition
ISBN: 9780134580999
Author: Elaine N. Marieb, Katja N. Hoehn
Publisher: PEARSON
expand_more
expand_more
format_list_bulleted
Question
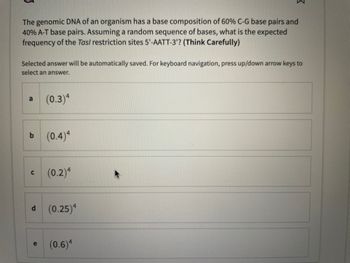
Transcribed Image Text:The genomic DNA of an organism has a base composition of 60% C-G base pairs and
40% A-T base pairs. Assuming a random sequence of bases, what is the expected
frequency of the Tas/ restriction sites 5'-AATT-3'? (Think Carefully)
Selected answer will be automatically saved. For keyboard navigation, press up/down arrow keys to
select an answer.
(0.3)4
a
b
C
d
e
(0.4)4
(0.2)4
(0.25)4
(0.6)4
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
This is a popular solution
Trending nowThis is a popular solution!
Step by stepSolved in 4 steps

Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biology and related others by exploring similar questions and additional content below.Similar questions
- The horizontal line represents the whole genome of Lambda DNA (a virus that infects a bacterial cell). The genome is 48,502 base pairs long. You are going to estimate the number and length in base pairs the fragments that result from cutting the lambda genome with three different enzymes (Eco R1, Hind III and Bam H1) Keep in mind that you are working with a linearized lambda DNA (has ends) because the DNA has been heated to 65oC and therefore, the cos site is not intact (not annealed)arrow_forwardYou have the following DNA sequence: 5'GGT ACG TTG GGG CTC CAT3' This sequence is transcribed and translated. If the G that is underlined changes to to a C the result will be - A) A nonsenese mutation B) A frameshift mutation C) A silent substitution D) A missense mutation You have the following DNA sequence: 5'GGT ACG TTG GGG CTC CAT3' This sequence is transcribed and translated. If the G that is underlined is deleted, then the result will be A) A nonsense mutation B) A frameshift mutation C) A silent substitution D) A missense mutatio If there are 3000 bases in the coding region of a gene, the gene will have A) 3000 amino acids B) 6000 amino acids C) 1000 amino acids D) 3000 codonsarrow_forward5' 3' O 60°C 5' O 95°C Primer 1 O 75°C ORF For your PCR reaction above, the primer set has a much higher AT (low GC) content than what would be considered normal. Due to this difference in AT content, the optimal annealing temperature for the primers is going to have to be altered from the "normal" temperature cycle: 95°C 60°C ⇒ 75°C Which of the temperatures will have to be altered to accommodate this difference the primer set? Primer 25¹ 3' 5' annealing witharrow_forward
- If a 1000 bp of DNA were inserted between the two restriction sites, how would the banding pattern on the gel differ from the one you drew in part a? (PART A WITH THE FIRST PART OF THE QUESTION IS ATTACHED)arrow_forwardGiven: BamHI, cleaves after the first G: 5’ G GATCC 3’ 3’ CCTAG G 5’ AND BclI cleaves after the first T: 5’ T GATCA 3’ 3’ ACTAG T 5’ THEN -- Given the DNA shown below: 5’ATTGAGGATCCGTAATGTGTCCTGATCACGCTCCACG3’ 3’TAACTCCTAGGCATTACACAGGACTAGTGCGAGGTGC5’ i) If this DNA was cut with BamHI, how many DNA fragments would you expect? ii) If the DNA shown above was cut with the enzyme BclI, how many DNA fragment would you expect?arrow_forwardThe sequences below indicated the 6bp recognition site for the restriction enzyme EcoRI. The lines indicate the sites where the enzyme will cut each strand. 1). write the sequence and structure of the two DNA pieces after the enzyme cuts (hydrogen bonds holding the strands together between the lines are broken after enzyme cuts) 2). indicate whether EcoRI generates blunt or sticky overhangs 5'- G I A A T T C - 3' 3' - C T T A A l G - 5'arrow_forward
- Given the DNA sequence of the restriction enzyme: gi|6329444|dbj|AB034757.1| Hynobius retardatus mRNA for larval beta-globin, complete cds GCAGAATCTGACTCAAGAAATCCCTCCTCACCCAACACCACCAGCAGCCATGGTTCACTGGACAGCAGAGGAGAAGGCAGCCATCAGCTCTGTGTGGAAGCAGGTGAACGTGGAGAGCGATGGACAGGAGGCCCTGGCCAGGTTGCTGATCGTCTACCCCTGGACCCAGAGATACTTCAGCTCTTTTGGGGACCTGTCGAGCCCAGCTGCCATTTGTGCCAACGCCAAGGTCCGTGCCCATGGCAAGAAGGTCCTGTCCGCCCTGGGAGCCGGCGCCAACCACCTGGATGACATCAAAGGCAACTTTGCTGATCTGAGCAAGCTTCACGCAGACACACTCCATGTGGACCCCAATAACTTCCTGCTCCTGGCAAACTGCCTGGTGATCGTCTTGGCCCGCAAGCTGGGAGCCGCCTTCAACCCTCAAGTCCATGCGGCCTGGGAGAAGTTCCTGGCCGTCTCCACCGCGGCTCTGTCCAGAAACTACCACTAGAGACTGGTCTTTGGGTTTAATTCTGTGAACGTCCCTGAGACAAATGATCTTTCAATGTGTAAACCTGTCATTACATCAATAAAGAGACATCTAACAAAAAAAAAAAAAAAAAAAAAAAAAA Identify two blunt-end cutters Identify two sticky-end cutters. For each, Provide the sequence of the Restriction enzyme, Highlight using a specific color where the DNA sequence where the restriction enzyme will cut the DNA Indicate the…arrow_forwardYou have two tubes, each with a pair of DNA fragments inside them. Tube #1 has fragments that are 500bp and 1000 bp in length. Tube #2 has fragments that are 7500bp and 8000bp in size. If you were to perform agarose gel electrophoresis and run the contents of each tube in two separate lanes on the same gel, what would you expect to see? O That the difference between the distances migrated by the two fragments in Tube #1 was much greater than the difference between the distances migrated by the two fragments in Tube #2. O That the difference between the distances migrated by the two fragments in Tube #1 was the same as difference between the distances migrated by the two fragments in Tube #2. O That the difference between the distances migrated by the two fragments in Tube #1 was much less than the difference between the distances migrated by the two fragments in Tube #2. O It is not possible to estimate what we would expect to see.arrow_forwardCan you please help with 1c please picture with 1 graph is for question 1a) picture with 4 graphs is for question 1b) 1a) E. coli DNA and binturong DNA are both 50% G-C. If you randomly shear E. coli DNA into 1000 bp fragments and put it through density gradient equilibrium centrifugation, you will find that all the DNA bands at the same place in the gradient, and if you graph the distribution of DNA fragments in the gradient you will get a single peak (see below). If you perform the same experiment with binturong DNA, you will find that a small fraction of the DNA fragments band separately in the gradient (at a different density) and give rise to a small "satellite" peak on a graph of the distribution of DNA fragments in the gradient (see below). Why do these two DNA samples give different results, when they're both 50% G-C? 1b) If you denatured the random 1000 bp fragments of binturong DNA that you produced in question 1a by heating them to 95ºC, and then cooled them down to 60ºC…arrow_forward
- 1) Prepare the following enzymatic reaction, present it in tabulated form. In a final volume of 30 ul, where buffer 4 (10 ml). How much volume of each reagent would be used and how much of water? Is there any problem? 2) The DNA pol 1 enzyme comes at a concentration of 50,000 U/ml. You have to prepare a 50 ug PCR reaction where you must use 0.05 U/ml reaction. You add 10 ul of PCR buffer, 2 ng of tempered DNA that is at a concentration of 0.5 ng/ul, primers (which are at 200 mM) so that each one remains at a concentration of 200 uM, Mg+2 that is 5 mM (10 X), enzyme and water. Present the table of all the reagents included in the reaction, the volumes of each one in ul. Present where the initial and final concentration of each reagent applies. Assume you have micropipettes for all values.arrow_forwardYou are studying a genome that has 42% G:C content and the remaining 58% of the genome is As and Ts. Assuming a random distribution of these bases what is the expected distance (ie there will be a cut once in every XX base pairs) between cut-sites for restriction enzyme that cuts at the sequence GAATTC? 256 3207 4168 4096arrow_forwardYou digest 4 uL of plasmid DNA that is 50 ng/uL concetration in a total volume of 20 uL. You run 10 uL of the digest on teh gel. You then do a DNA purification protocol with a Zippy prep on the remaining digested DNA. You elute the DNA in a 25 uL. A 2 uL ssample has the concentration of 2 ng/uL. What is the DNA yield?arrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 Human Anatomy & Physiology (11th Edition)BiologyISBN:9780134580999Author:Elaine N. Marieb, Katja N. HoehnPublisher:PEARSON
Human Anatomy & Physiology (11th Edition)BiologyISBN:9780134580999Author:Elaine N. Marieb, Katja N. HoehnPublisher:PEARSON Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax
Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax Anatomy & PhysiologyBiologyISBN:9781259398629Author:McKinley, Michael P., O'loughlin, Valerie Dean, Bidle, Theresa StouterPublisher:Mcgraw Hill Education,
Anatomy & PhysiologyBiologyISBN:9781259398629Author:McKinley, Michael P., O'loughlin, Valerie Dean, Bidle, Theresa StouterPublisher:Mcgraw Hill Education, Molecular Biology of the Cell (Sixth Edition)BiologyISBN:9780815344322Author:Bruce Alberts, Alexander D. Johnson, Julian Lewis, David Morgan, Martin Raff, Keith Roberts, Peter WalterPublisher:W. W. Norton & Company
Molecular Biology of the Cell (Sixth Edition)BiologyISBN:9780815344322Author:Bruce Alberts, Alexander D. Johnson, Julian Lewis, David Morgan, Martin Raff, Keith Roberts, Peter WalterPublisher:W. W. Norton & Company Laboratory Manual For Human Anatomy & PhysiologyBiologyISBN:9781260159363Author:Martin, Terry R., Prentice-craver, CynthiaPublisher:McGraw-Hill Publishing Co.
Laboratory Manual For Human Anatomy & PhysiologyBiologyISBN:9781260159363Author:Martin, Terry R., Prentice-craver, CynthiaPublisher:McGraw-Hill Publishing Co. Inquiry Into Life (16th Edition)BiologyISBN:9781260231700Author:Sylvia S. Mader, Michael WindelspechtPublisher:McGraw Hill Education
Inquiry Into Life (16th Edition)BiologyISBN:9781260231700Author:Sylvia S. Mader, Michael WindelspechtPublisher:McGraw Hill Education

Human Anatomy & Physiology (11th Edition)
Biology
ISBN:9780134580999
Author:Elaine N. Marieb, Katja N. Hoehn
Publisher:PEARSON

Biology 2e
Biology
ISBN:9781947172517
Author:Matthew Douglas, Jung Choi, Mary Ann Clark
Publisher:OpenStax

Anatomy & Physiology
Biology
ISBN:9781259398629
Author:McKinley, Michael P., O'loughlin, Valerie Dean, Bidle, Theresa Stouter
Publisher:Mcgraw Hill Education,

Molecular Biology of the Cell (Sixth Edition)
Biology
ISBN:9780815344322
Author:Bruce Alberts, Alexander D. Johnson, Julian Lewis, David Morgan, Martin Raff, Keith Roberts, Peter Walter
Publisher:W. W. Norton & Company

Laboratory Manual For Human Anatomy & Physiology
Biology
ISBN:9781260159363
Author:Martin, Terry R., Prentice-craver, Cynthia
Publisher:McGraw-Hill Publishing Co.

Inquiry Into Life (16th Edition)
Biology
ISBN:9781260231700
Author:Sylvia S. Mader, Michael Windelspecht
Publisher:McGraw Hill Education