
Human Anatomy & Physiology (11th Edition)
11th Edition
ISBN: 9780134580999
Author: Elaine N. Marieb, Katja N. Hoehn
Publisher: PEARSON
expand_more
expand_more
format_list_bulleted
Concept explainers
Question
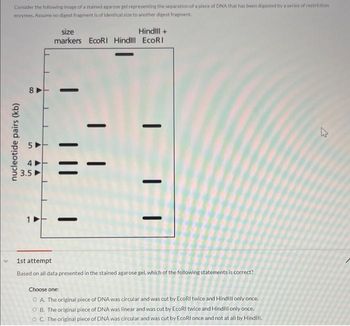
Transcribed Image Text:Consider the following image of a stained agarose gel representing the separation of a plece of DNA that has been digested by a series of restriction
enzymes. Assume no digest fragment is of identical size to another digest fragment.
nucleotide pairs (kb)
8
S
45
3.5
AA
size
Hindill +
markers EcoRI Hindill EcoRI
-
1st attempt
Based on all data presented in the stained agarose gel, which of the following statements is correct?
Choose one:
OA. The original piece of DNA was circular and was cut by EcoRI twice and Hindill only once.
OB. The original piece of DNA was linear and was cut by EcoRI twice and Hindill only once.
OC. The original piece of DNA was circular and was cut by EcoRI once and not at all by Hind!!!.
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
This is a popular solution
Trending nowThis is a popular solution!
Step by stepSolved in 4 steps

Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biology and related others by exploring similar questions and additional content below.Similar questions
- DNA samples from four individuals were cleaved with the same MW restriction endonuclease. The DNA fragments were separated by gel clectrophoresis, transferred to a membrane, and hybridized with a 12 kb 10 kb DNA probe complementary to a region between sites C and D (see 8 kb hybridization line). The image of the southern blot shows the labeled DNA bands and 6 kb molecular weight (MW) markers. The lane labels I, II, III, and IV -5 kb correspond to individuals I, II, III, and IV. Assume that fragments such as C-D and C-E are clearly resolved in this gel system. Fragment sizes are as given: A-B is 4 kb, B-C is I kb, C-D is 5 kb, and D-E is 650 bp. Individual I has five cleavage sites (A, B, C, D, and E) for the restriction endonuclease. DNA homologous to probe Which individual has at least one point mutation that eliminates restriction site C only? O II IV cannot be determined II Which individual has at least one point mutation that climinates restriction sites B and C? III O IV cannot be…arrow_forwardHow many fragments and how big each fragment should be after digesting the plasmid with BamHI? How many fragments and how big each fragment should be after digesting the plasmid with PvuI? How many fragments and how big each fragment should be after digesting with both enzymes? Show your calculation/explanation.arrow_forwardHow much 340bp insert DNA should be added to a ligation in which 50ng of 4.3kb vector will be used? The desired vector: insert molar ratio needs to be 1:3.arrow_forward
- 1) Prepare the following enzymatic reaction, present it in tabulated form. In a final volume of 30 ul, where buffer 4 (10 ml). How much volume of each reagent would be used and how much of water? Is there any problem? 2) The DNA pol 1 enzyme comes at a concentration of 50,000 U/ml. You have to prepare a 50 ug PCR reaction where you must use 0.05 U/ml reaction. You add 10 ul of PCR buffer, 2 ng of tempered DNA that is at a concentration of 0.5 ng/ul, primers (which are at 200 mM) so that each one remains at a concentration of 200 uM, Mg+2 that is 5 mM (10 X), enzyme and water. Present the table of all the reagents included in the reaction, the volumes of each one in ul. Present where the initial and final concentration of each reagent applies. Assume you have micropipettes for all values.arrow_forwardAssume complete digestion of a single linear piece of DNA digested (cut) with EcoRI and Hindill. Assume complete digestion of the plasmid digested with Eco and HindIll. What comes out of the plasmid when you digest with EcoRl and Hindill? What could ligate into the plasmid during ligation! Need help answering these questionsarrow_forwardYou are studying a genome that has 42% G:C content and the remaining 58% of the genome is As and Ts. Assuming a random distribution of these bases what is the expected distance (ie there will be a cut once in every XX base pairs) between cut-sites for restriction enzyme that cuts at the sequence GAATTC? 256 3207 4168 4096arrow_forward
- Use the gel to answer the following questions. You will be constructing a map of the plasmid, pDiddy. What is the smallest fragment size that the NcoI/EcoRI double digest produces?arrow_forwardDescribe the process for shotgun sequencing of a genome. Practice aligning the two sets of sequenced fragments below, to determine the order of the fragments and the complete sequence.arrow_forwardA 2.0kb bacterial plasmid ‘BS1030’ is digested with the restriction endonuclease Sau3A; the plasmid map is depicted in the diagram below and the Sau3A (S) restriction sites are indicated. Which of the following DNA fragments do you expect to see on an agarose gel when you run Sau3A-digested plasmid ‘BS1030’ DNA? a. 250 bp, 450 bp, 550 bp, 1.1 kb, 1.5 kb and 2.0 kb b. 2.0kb c. 250 bp, 400 bp, 450 bp, 500 bp and 550 bp d. 100 bp, 200 bp, 250 bp, 400 bp, 500 bp and 550 bparrow_forward
- Assume that a plasmid is 4700 base pairs in length and has restriction sites for a given restriction enzyme at the following locations: 800, 1400, 2900, and 3600. List the fragments by size that are ! expected when the plasmid is fully digested the restriction enzyme.arrow_forwardAll of the following are performed during restriction fragment length polymorphism analysis. 1. splitting of double-stranded into single-stranded DNA 2. gel electrophoresis 3. autoradiography 4. immersion in radioactive probes 5. digestion of DNA with restriction endonucleases 6. use of a positive charge to transfer single-stranded DNA from a gel to a membrane. The correct sequence of these operations is whatarrow_forwardPlease explain in detail....arrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 Human Anatomy & Physiology (11th Edition)BiologyISBN:9780134580999Author:Elaine N. Marieb, Katja N. HoehnPublisher:PEARSON
Human Anatomy & Physiology (11th Edition)BiologyISBN:9780134580999Author:Elaine N. Marieb, Katja N. HoehnPublisher:PEARSON Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax
Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax Anatomy & PhysiologyBiologyISBN:9781259398629Author:McKinley, Michael P., O'loughlin, Valerie Dean, Bidle, Theresa StouterPublisher:Mcgraw Hill Education,
Anatomy & PhysiologyBiologyISBN:9781259398629Author:McKinley, Michael P., O'loughlin, Valerie Dean, Bidle, Theresa StouterPublisher:Mcgraw Hill Education, Molecular Biology of the Cell (Sixth Edition)BiologyISBN:9780815344322Author:Bruce Alberts, Alexander D. Johnson, Julian Lewis, David Morgan, Martin Raff, Keith Roberts, Peter WalterPublisher:W. W. Norton & Company
Molecular Biology of the Cell (Sixth Edition)BiologyISBN:9780815344322Author:Bruce Alberts, Alexander D. Johnson, Julian Lewis, David Morgan, Martin Raff, Keith Roberts, Peter WalterPublisher:W. W. Norton & Company Laboratory Manual For Human Anatomy & PhysiologyBiologyISBN:9781260159363Author:Martin, Terry R., Prentice-craver, CynthiaPublisher:McGraw-Hill Publishing Co.
Laboratory Manual For Human Anatomy & PhysiologyBiologyISBN:9781260159363Author:Martin, Terry R., Prentice-craver, CynthiaPublisher:McGraw-Hill Publishing Co. Inquiry Into Life (16th Edition)BiologyISBN:9781260231700Author:Sylvia S. Mader, Michael WindelspechtPublisher:McGraw Hill Education
Inquiry Into Life (16th Edition)BiologyISBN:9781260231700Author:Sylvia S. Mader, Michael WindelspechtPublisher:McGraw Hill Education

Human Anatomy & Physiology (11th Edition)
Biology
ISBN:9780134580999
Author:Elaine N. Marieb, Katja N. Hoehn
Publisher:PEARSON

Biology 2e
Biology
ISBN:9781947172517
Author:Matthew Douglas, Jung Choi, Mary Ann Clark
Publisher:OpenStax

Anatomy & Physiology
Biology
ISBN:9781259398629
Author:McKinley, Michael P., O'loughlin, Valerie Dean, Bidle, Theresa Stouter
Publisher:Mcgraw Hill Education,

Molecular Biology of the Cell (Sixth Edition)
Biology
ISBN:9780815344322
Author:Bruce Alberts, Alexander D. Johnson, Julian Lewis, David Morgan, Martin Raff, Keith Roberts, Peter Walter
Publisher:W. W. Norton & Company

Laboratory Manual For Human Anatomy & Physiology
Biology
ISBN:9781260159363
Author:Martin, Terry R., Prentice-craver, Cynthia
Publisher:McGraw-Hill Publishing Co.

Inquiry Into Life (16th Edition)
Biology
ISBN:9781260231700
Author:Sylvia S. Mader, Michael Windelspecht
Publisher:McGraw Hill Education