
Concept explainers
Can you please help with 1c please
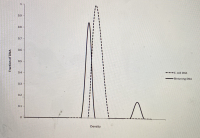
picture with 1 graph is for question 1a)
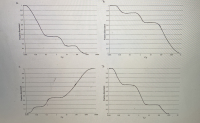
picture with 4 graphs is for question 1b)
1a) E. coli DNA and binturong DNA are both 50% G-C. If you randomly shear E. coli DNA into 1000 bp fragments and put it through density gradient equilibrium centrifugation, you will find that all the DNA bands at the same place in the gradient, and if you graph the distribution of DNA fragments in the gradient you will get a single peak (see below). If you perform the same experiment with binturong DNA, you will find that a small fraction of the DNA fragments band separately in the gradient (at a different density) and give rise to a small "satellite" peak on a graph of the distribution of DNA fragments in the gradient (see below). Why do these two DNA samples give different results, when they're both 50% G-C?
1b) If you denatured the random 1000 bp fragments of binturong DNA that you produced in question 1a by heating them to 95ºC, and then cooled them down to 60ºC and allowed them to reanneal, you would find that approximately 15% of the DNA fragments renatured very rapidly, another 20% of the DNA fragments renatured moderately rapidly, and the remaining DNA fragments renatured relatively slowly. Which of the graphs shown below is a C0t curve showing these results?
1c) What fraction of the binturong genomic DNA would be considered to be "highly repetitive" DNA, based on this data?
Deoxyribonucleic acidi an atom made out of two polynucleotide chains that curl around one another to frame a twofold helix conveying hereditary directions for the turn of events, working, development and multiplication of every single known creature and numerous infections. DNA and ribonucleic acid are nucleic acids.
Step by stepSolved in 2 steps

- Why was it necessary to mash the strawberries extensively with your hands? (hint: you wouldn’t have to do this step with an animal cells). Why do we treat the cells with soap when conducting DNA extraction? And why do we add salt when doing the DNA extraction?arrow_forwardIn hybridizing microarrays, we use a solution that contains {3X SSC, 0.5% SDS, 5ug/ml salmon sperm DNA, 5% glycerol}. Describe how to make 100ul of this solution using stocks of 20X SSC, 10% SDS, and 10 mg/ml salmon sperm DNA, sheared, in water. Note that Molecular Biology Grade H2O must be used in making up this solution.arrow_forwardBiology Questionarrow_forward
- You have two tubes, each with a pair of DNA fragments inside them. Tube #1 has fragments that are 500bp and 1000 bp in length. Tube #2 has fragments that are 7500bp and 8000bp in size. If you were to perform agarose gel electrophoresis and run the contents of each tube in two separate lanes on the same gel, what would you expect to see? O That the difference between the distances migrated by the two fragments in Tube #1 was much greater than the difference between the distances migrated by the two fragments in Tube #2. O That the difference between the distances migrated by the two fragments in Tube #1 was the same as difference between the distances migrated by the two fragments in Tube #2. O That the difference between the distances migrated by the two fragments in Tube #1 was much less than the difference between the distances migrated by the two fragments in Tube #2. O It is not possible to estimate what we would expect to see.arrow_forwardYou used agarose gel electrophoresis to separate DNA fragments of different size and the experiment worked well. However, you wanted to re run the experiment but this time you made the gel with a higher percentage of Agarose. How might this affect your results compared to the first run? a. There would be no difference between the runs since it is the current, not the agarose that causes migration. b. The higher concentration of agarose would cause the DNA to break apart. c. You can't predict how the concetration of agarose would affect migration. d. The DNA fragments would migrate further down the gel than they did the first time. e. The DNA fragments wouldn't migrate as far down the gel as they did the first time.arrow_forwardCan you please help with 1c please picture with 1 graph is for question 1a) picture with 4 graphs is for question 1b) 1a) E. coli DNA and binturong DNA are both 50% G-C. If you randomly shear E. coli DNA into 1000 bp fragments and put it through density gradient equilibrium centrifugation, you will find that all the DNA bands at the same place in the gradient, and if you graph the distribution of DNA fragments in the gradient you will get a single peak (see below). If you perform the same experiment with binturong DNA, you will find that a small fraction of the DNA fragments band separately in the gradient (at a different density) and give rise to a small "satellite" peak on a graph of the distribution of DNA fragments in the gradient (see below). Why do these two DNA samples give different results, when they're both 50% G-C? 1b) If you denatured the random 1000 bp fragments of binturong DNA that you produced in question 1a by heating them to 95ºC, and then cooled them down to 60ºC…arrow_forward
- 1) Prepare the following enzymatic reaction, present it in tabulated form. In a final volume of 30 ul, where buffer 4 (10 ml). How much volume of each reagent would be used and how much of water? Is there any problem? 2) The DNA pol 1 enzyme comes at a concentration of 50,000 U/ml. You have to prepare a 50 ug PCR reaction where you must use 0.05 U/ml reaction. You add 10 ul of PCR buffer, 2 ng of tempered DNA that is at a concentration of 0.5 ng/ul, primers (which are at 200 mM) so that each one remains at a concentration of 200 uM, Mg+2 that is 5 mM (10 X), enzyme and water. Present the table of all the reagents included in the reaction, the volumes of each one in ul. Present where the initial and final concentration of each reagent applies. Assume you have micropipettes for all values.arrow_forwardSecond, Question 3arrow_forwardA 2.0kb bacterial plasmid ‘BS1030’ is digested with the restriction endonuclease Sau3A; the plasmid map is depicted in the diagram below and the Sau3A (S) restriction sites are indicated. Which of the following DNA fragments do you expect to see on an agarose gel when you run Sau3A-digested plasmid ‘BS1030’ DNA? a. 250 bp, 450 bp, 550 bp, 1.1 kb, 1.5 kb and 2.0 kb b. 2.0kb c. 250 bp, 400 bp, 450 bp, 500 bp and 550 bp d. 100 bp, 200 bp, 250 bp, 400 bp, 500 bp and 550 bparrow_forward
- Assuming that the mean size of the human sau3AI partially digested fragments clones is 3kb, calculate the proportion of the human genome represented in all libraries prepared by the class. Use a value of 3,000,000kb for the size of the human genome. You also need to know that the whole 'class' generated 3,514 white colonies on plate 2.arrow_forwardWhich one is the correct one?arrow_forwardAnswer part C of the following: A) E. coli DNA and binturong DNA are both 50% G-C. If you randomly shear E. coli DNA into 1000 bp fragments and put it through density gradient equilibrium centrifugation, you will find that all the DNA bands at the same place in the gradient, and if you graph the distribution of DNA fragments in the gradient you will get a single peak (see below). If you perform the same experiment with binturong DNA, you will find that a small fraction of the DNA fragments band separately in the gradient (at a different density) and give rise to a small "satellite" peak on a graph of the distribution of DNA fragments in the gradient (see below). Why do these two DNA samples give different results, when they're both 50% G-C? See graph B. B) If you denatured the random 1000 bp fragments of binturong DNA that you produced in question 1a by heating them to 95ºC, and then cooled them down to 60ºC and allowed them to reanneal, you would find that approximately 15% of the…arrow_forward
 Human Anatomy & Physiology (11th Edition)BiologyISBN:9780134580999Author:Elaine N. Marieb, Katja N. HoehnPublisher:PEARSON
Human Anatomy & Physiology (11th Edition)BiologyISBN:9780134580999Author:Elaine N. Marieb, Katja N. HoehnPublisher:PEARSON Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax
Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax Anatomy & PhysiologyBiologyISBN:9781259398629Author:McKinley, Michael P., O'loughlin, Valerie Dean, Bidle, Theresa StouterPublisher:Mcgraw Hill Education,
Anatomy & PhysiologyBiologyISBN:9781259398629Author:McKinley, Michael P., O'loughlin, Valerie Dean, Bidle, Theresa StouterPublisher:Mcgraw Hill Education, Molecular Biology of the Cell (Sixth Edition)BiologyISBN:9780815344322Author:Bruce Alberts, Alexander D. Johnson, Julian Lewis, David Morgan, Martin Raff, Keith Roberts, Peter WalterPublisher:W. W. Norton & Company
Molecular Biology of the Cell (Sixth Edition)BiologyISBN:9780815344322Author:Bruce Alberts, Alexander D. Johnson, Julian Lewis, David Morgan, Martin Raff, Keith Roberts, Peter WalterPublisher:W. W. Norton & Company Laboratory Manual For Human Anatomy & PhysiologyBiologyISBN:9781260159363Author:Martin, Terry R., Prentice-craver, CynthiaPublisher:McGraw-Hill Publishing Co.
Laboratory Manual For Human Anatomy & PhysiologyBiologyISBN:9781260159363Author:Martin, Terry R., Prentice-craver, CynthiaPublisher:McGraw-Hill Publishing Co. Inquiry Into Life (16th Edition)BiologyISBN:9781260231700Author:Sylvia S. Mader, Michael WindelspechtPublisher:McGraw Hill Education
Inquiry Into Life (16th Edition)BiologyISBN:9781260231700Author:Sylvia S. Mader, Michael WindelspechtPublisher:McGraw Hill Education





