
Concept explainers
To further analyze the CRABS CLAW gene (see Problems

What restriction digest would help resolve any ambiguity in the map?
You have isolated a genomic clone with an EcoRI fragment of
Does this tell you anything about where the CRABS CLAW gene is located within the 11-kb genomic clone?
Restriction enzyme sites within a cDNA clone are often also found in the genomic sequence. Can you think of a reason why occasionally this is not the case? What about the converse: Are restriction enzyme sites in a genomic clone always in a cDNA clone of the same gene?
You have identified a
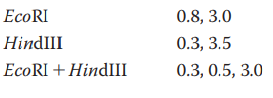
Want to see the full answer?
Check out a sample textbook solution
Chapter 15 Solutions
Genetic Analysis: An Integrated Approach (3rd Edition)
- Below is a diagram of the vector you are planning to use. You identify four restriction enzyme recognition sites in the vector as indicated in the diagram below. The distance between the restriction sites are indicated by numbers (in kilo base pairs). ori ВатHI ВатHI 2 ЕcoRI Kpnl If you digest the vector with a combination of EcoRI and BamHI and run the resulting DNA fragments on a gel, which lane would represent the expected result? (Lane 1 contains the DNA size marker.) А В с (kbp) 8 Promoterarrow_forwardWhen the restriction endonuclease EcoRI is used to digest a 10 kb DNA fragment, it produces 4 kb and 6 kb-sized fragments. Digesting the 10 kb fragment with BamHI yields three fragments, each ranging in size from one to three and a half kilobytes. Four pieces of 0.5, 1, 3 and 5.5 kb are formed after using both enzymes. Create a restriction map for this 10 kb piece of DNA using the information you have collected. Make a note of where the two enzymes cut, as well as the distances between the enzymes.arrow_forwardA cloned Bam HI DNA fragment, 3.0 kb long, was isolated in preparation for sequence determination. First, a map of restriction endonuclease cleavage sites had to be prepared. Cleavage by each of the indicated enzymes generated fragments of the sizes shown on the three agarose electrophoresis gel figures below; the gels are running from top to bottom. On the diagram below those, show the relative positions of EcoR I and Hpa II cleavage sites and the distances (in kb) between them. Explain your reasoning – is your placement of the cleavage sites the only one that will work?arrow_forward
- Many restriction endonuclease recognition sequences are palindromes. What are palindromes? Are the recognition sequences for AatII and DraI palindromes? Some restriction endonucleases produce blunt-ended pieces of DNA, while other produce DNA fragments with sticky ends. What is the difference? What type of ends do AatII and DraI produce?arrow_forwardA 10 kb DNA fragment digested with the restriction endonuclease EcoRI yields fragments of 4 kb and 6 kb. When the 10 kb fragment is digested with BamHI, three fragments of 1, 3.5 and 5.5 kb are generated. Digestion with both enzymes yields four fragments of 0.5, 1, 3 and 5.5 kb. Draw the restriction map for the 10 kb fragment based on the data. Label the cut sites for the two enzymes, and the lengths between the cut sites.arrow_forwardGenomic DNA from a family where sickle-cell disease is known to be hereditary, is digested with the restriction enzyme MstII and run in a Southern Blot. The blot is hybridised with two different 0.6 kb probes, both probes (indicated in red in the diagram below) are specific for the β-globin gene (indicated as grey arrow on the diagram below). The normal wild-type βA allele contains an MstII restriction site indicated with the asterisk (*) in the diagram below; in the mutated sickle-cell βS allele this restriction site has been lost. What size bands would you expect to see on the Southern blots using probe 1 and probe 2 for an individual with sickle cell disease (have 2 βS alleles)? Probe 1 Probe 2 (a) 0.6kb 0.6kb and 1.2kb (b) 0.6kb and 1.8kb 0.6kb, 1.2kb and 1.8kb (c) 1.2kb 0.6kb (d) 1.8kb 1.8kb a. (a) b. (b) c. (c) d. (d)arrow_forward
- Transcriptome analysis involves two separate methodologies: gene expression and RNA seq analyses. The 10 items below are a scrambled listing of the steps used in the two procedures. Identify the steps involved in RNA seq from the list below. Use the numbers in the list to refer to each step. Once the steps for RNA seq have been identified, write the steps in the order in which they are performed during the experiment. (1) DNA sequencing (2) Allow for hybridization and wash excess cRNA. (3) Mix labeled cRNA with array chip. (4) PCR amplification (5) Measure fluorescence intensity to determine abundance of transcripts. (6) Add labeled cRNA at each microarray location. (7) Map cDNA sequences to the genome of the organism to determine identity and abundance of transcripts. (8) mRNA isolation from cells (9) Prepare fluorescently labeled cRNA probes (10) cDNA synthesisarrow_forwardKnowing that you are using HindIII and EcoRI to cut your plasmids, and that those two enzymes cut within the MCS, use the map of pUC19 provided below to compute: What will be the sizes of the 2 restriction fragments if NO insert is present in pUC19? What will be the sizes of the 2 restriction fragments if the approximately 317 bp RT-PCR product (insert from WT satC dimer) was ligated successfully into the SmaI site? What will be the sizes of the 2 restriction fragments if TWO approximately 317 bp RT-PCR products (2 ligated inserts from WT satC dimer) were ligated into the SmaI site?arrow_forwardThe figure below shows the recognition sequences and cleavage positions of three restriction enzymes.You plan to ligate DNA from two different sources. The target DNA is digested with BamHI,and the insert DNA is digested with BglII, and the resulting fragments mixed and incubatedwith DNA ligase. a) Write out the sequence (in double-stranded format) of the longest insert fragment that will result after BglII digestion, ensure the nature of the overhangs is clear.b) Write out the sequence (in double-stranded format) of the ligation product, with the insert fragment joined into the BamHI site of the target DNA. Use black for target sequences, and blue for insert sequences. c) Assume the ligation reaction was successful and you have generated a recombinant DNAmolecule. Which of the three enzymes listed above can be used to excise the insert DNAfrom the target? Motivate your answer.arrow_forward
- For Pet41 (choose Pet41 a, b, or c as provided in the image) how would you design the primers (forward and reverse) for the following gene of interest and what restriction enzymes would be used (as shown in the image)? Be sure to explain and elaborate on why selected and how. Gene of Interest: atgggc gacaaaggga 241 cccgagtgtt caagaaggcc agtccaaatg gaaagctcac cgtctacctg ggaaagcggg 301 actttgtgga ccacatcgac ctcgtggacc ctgtggatgg tgtggtcctg gtggatcctg 361 agtatctcaa agagcggaga gtctatgtga cgctgacctg cgccttccgc tatggccggg 421 aggacctgga tgtcctgggc ctgacctttc gcaaggacct gtttgtggcc aacgtacagt 481 cgttcccacc ggcccccgag gacaagaagc ccctgacgcg gctgcaggaa cgcctcatca 541 agaagctggg cgagcacgct taccctttca cctttgagat ccctccaaac cttccatgtt 601 ctgtgacact gcagccgggg cccgaagaca cggggaaggc ttgcggtgtg gactatgaag 661 tcaaagcctt ctgcgcggag aatttggagg agaagatcca caagcggaat tctgtgcgtc 721 tggtcatccg gaaggttcag tatgccccag agaggcctgg cccccagccc acagccgaga 781 ccaccaggca gttcctcatg tcggacaagc ccttgcacct…arrow_forwardYou are trying to clone a gene. You have successfully isolated it from the genomic DNA of an organism using the Hindlll restriction enzyme. You then take a plasmid with a single EcoRI restriction site and cleave it with EcoRI. You combine these two fragments and treat them with DNA ligase. Answer the two questions below. a.(2 points Does the cloning reaction succeed as described? If so, what is the product obtained? b. Explain your answer above.arrow_forward1) You wish to make a restriction map of a 17.0 kb linear fragment. You digest the fragment with Sbf1, Pst1, and a mixture of Sbf1 and Pst1. From these results, you obtain these fragments following agarose gel electrophoresis of the three samples: Sbf1: 7kb and 10kb Pstl: 3kb, 6kb and 8kb Sbf1 and Pstl: 1kb, 2kb, 6kb and 8kb Question: In which of the Pstl fragments is the Sbfl site located? 8kb O 6kb 3kb O None of the abovearrow_forward
 Human Anatomy & Physiology (11th Edition)BiologyISBN:9780134580999Author:Elaine N. Marieb, Katja N. HoehnPublisher:PEARSON
Human Anatomy & Physiology (11th Edition)BiologyISBN:9780134580999Author:Elaine N. Marieb, Katja N. HoehnPublisher:PEARSON Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax
Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax Anatomy & PhysiologyBiologyISBN:9781259398629Author:McKinley, Michael P., O'loughlin, Valerie Dean, Bidle, Theresa StouterPublisher:Mcgraw Hill Education,
Anatomy & PhysiologyBiologyISBN:9781259398629Author:McKinley, Michael P., O'loughlin, Valerie Dean, Bidle, Theresa StouterPublisher:Mcgraw Hill Education, Molecular Biology of the Cell (Sixth Edition)BiologyISBN:9780815344322Author:Bruce Alberts, Alexander D. Johnson, Julian Lewis, David Morgan, Martin Raff, Keith Roberts, Peter WalterPublisher:W. W. Norton & Company
Molecular Biology of the Cell (Sixth Edition)BiologyISBN:9780815344322Author:Bruce Alberts, Alexander D. Johnson, Julian Lewis, David Morgan, Martin Raff, Keith Roberts, Peter WalterPublisher:W. W. Norton & Company Laboratory Manual For Human Anatomy & PhysiologyBiologyISBN:9781260159363Author:Martin, Terry R., Prentice-craver, CynthiaPublisher:McGraw-Hill Publishing Co.
Laboratory Manual For Human Anatomy & PhysiologyBiologyISBN:9781260159363Author:Martin, Terry R., Prentice-craver, CynthiaPublisher:McGraw-Hill Publishing Co. Inquiry Into Life (16th Edition)BiologyISBN:9781260231700Author:Sylvia S. Mader, Michael WindelspechtPublisher:McGraw Hill Education
Inquiry Into Life (16th Edition)BiologyISBN:9781260231700Author:Sylvia S. Mader, Michael WindelspechtPublisher:McGraw Hill Education





