
Concept explainers
Alkaptonuria is a human autosomal recessive disorder caused by mutation of the HAO gene that encodes the enzyme homogentisic acid oxidase. Restriction mapping
of the HAO gene region reveals four BamHI restriction sites
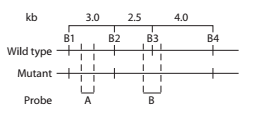
DNA samples taken from a mother (M), father (F), and two children
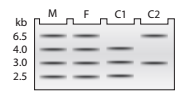
a. Using A to represent the
b. In a separate figure, draw the autoradiograph patterns for all the genotypes that could be found in children of this couple.
c. Explain how the DNA sequence change results in a
Want to see the full answer?
Check out a sample textbook solution
Chapter 11 Solutions
Genetic Analysis: An Integrated Approach (3rd Edition)
- What are Clathrin coated vesicles and what is their function?arrow_forwardHow is a protein destined for the Endoplasmic Reticulum (ER), imported into the ER? Be concise.arrow_forwardFind out about the organisations and the movements aimed at the conservation of our natural resources. Eg Chipko movement and Greenpeace. Make a project report on such an organisation.arrow_forward
- What are biofertilizers and mention the significancearrow_forwardPCBs and River Otters: Otters in Washington State’s Green-Duwamish River have high levels of polychlorinated biphenyls (PCBs) in their livers. PCBs can bind to the estrogen receptors in animals and disrupt the endocrine system of these otters. The PCBs seem to increase the estrogen to androgen ratio, skewing the ratio toward too much estrogen. How would increased estrogen affect the river otter population? Based on your reading of the materials in this unit, what factors can affect fertility in humans? Explain how each of the factors affecting human fertility that you described can disrupt the human endocrine system to affect reproduction.arrow_forwardOther than oil and alcohol, are there other liquids you could compare to water (that are liquid at room temperature)? How is water unique compared to these other liquids? What follow-up experiment would you like to do, and how would you relate it to your life?arrow_forward
- Selection of Traits What adaptations do scavengers have for locating and feeding on prey? What adaptations do predators have for capturing and consuming prey?arrow_forwardCompetition Between Species What natural processes limit populations from growing too large? What are some resources organisms can compete over in their natural habitat?arrow_forwardSpecies Interactions Explain how predators, prey and scavengers interact. Explain whether predators and scavengers are necessary or beneficial for an ecosystem.arrow_forward
- magine that you are conducting research on fruit type and seed dispersal. You submitted a paper to a peer-reviewed journal that addresses the factors that impact fruit type and seed dispersal mechanisms in plants of Central America. The editor of the journal communicates that your paper may be published if you make ‘minor revisions’ to the document. Describe two characteristics that you would expect in seeds that are dispersed by the wind. Contrast this with what you would expect for seeds that are gathered, buried or eaten by animals, and explain why they are different. (Editor’s note: Providing this information in your discussion will help readers to consider the significance of the research).arrow_forwardWhat is the difference between Uniporters, Symporters and Antiporters? Which of these are examples of active transport?arrow_forwardWhat are coupled transporters?arrow_forward
 Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage LearningCase Studies In Health Information ManagementBiologyISBN:9781337676908Author:SCHNERINGPublisher:Cengage
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage LearningCase Studies In Health Information ManagementBiologyISBN:9781337676908Author:SCHNERINGPublisher:Cengage Human Biology (MindTap Course List)BiologyISBN:9781305112100Author:Cecie Starr, Beverly McMillanPublisher:Cengage Learning
Human Biology (MindTap Course List)BiologyISBN:9781305112100Author:Cecie Starr, Beverly McMillanPublisher:Cengage Learning Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning
Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax
Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax





