
Biochemistry
9th Edition
ISBN: 9781319114671
Author: Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.
Publisher: W. H. Freeman
expand_more
expand_more
format_list_bulleted
Concept explainers
Question

Transcribed Image Text:Using the TABLE shown, identify which proteins would elute together in a gel filtration experiment.
1 & 2 and 4 & 5
1, 2 & 4
2 & 3
1 & 2
4 & 5
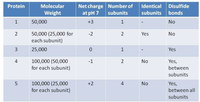
Transcribed Image Text:Net charge Number of Identical Disulfide
subunits
Protein
Molecular
Weight
at pH 7
subunits bonds
1
50,000
+3
1
No
50,000 (25,000 for
each subunit)
2
-2
2
Yes
No
25,000
Yes
100,000 (50,000
for each subunit)
4
-1
2
No
Yes,
between
subunits
100,000 (25,000
for each subunit)
5
+2
4
No
Yes,
between all
subunits
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
This is a popular solution
Trending nowThis is a popular solution!
Step by stepSolved in 3 steps

Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biochemistry and related others by exploring similar questions and additional content below.Similar questions
- Compare and contrast the following protein characterization techniques in terms of the principles governing their functions. electrosrapy ionization mass spectrometry vs polyacrylamide gel electrophoresisarrow_forwardFive dialysis bags, impermeable to sucrose, were filled with various concentrations of sucrose and then placed in separate beakers containing an initial concentration of 0.6 M sucrose solution. At 10 minute intervals, the bags were weighed and the percent change in mass of each bag was graphed. Which line represents the bag that contained a solution isotonic to the 0.6 M solution at the beginning of the experiment? Which line represents the bag with the highest initial concentration of sucrose? Which line or lines represent bags that contain a solution that is still hypertonic at the end of 60 minutes? What is the best explanation for the shape of line e. after 50 minutes?arrow_forwardExplain how to conduct a Kirby-Bauer disk diffusion test. Include all of the steps involved.arrow_forward
- You perform a Bradford assay. You obtain the absorbance values listed below from the BSA samples; your protein sample yields an absorbance of 1.3; what is the protein concentration of your sample? How did you determine that? BSA (ug/ml) Absorbance @ 595 nm. 25 0.15 50 0.30 75 0.45 100 0.60 150 0.90 200 1.25arrow_forwardIn this problem, you will design a filter system for a flow cytometer. The four fluorescent dyes are FITC,PE, PE-TR,and PE-Cy5. Their emisison spectra are provided below. Detectors are arranged as shown in thefigure below. The FITC signal is detected from Filter 1, the PE signal from Filter 2, the PE-TR signal fromFilter 3 and the PE-Cy 5 signal from Filter 4. Part 4.1Provide a rationale for the order in which the filters are arranged with respect to the unfiltered signal. Part 4.2You are given 9 candidates from which you have to choose 4 for the filters in the diagram above. Choose4 and justify your selection.# Filter type WavelengthA Long pass 650 nmB Short pass 530 nmC Long pass 490 nmD Band pass 550-590 nmE Long pass 560 nmF Short pass 640 nmG Long pass 620 nmH Band pass 620-640 nmI Short pass 590 nmarrow_forwardWhich fractions measured from your gel filtration experiment had the most and least amount of protein detected? Which fraction likely contained the most myoglobin based off molecular weight and protein presence? Which fraction likely contained the most egg albumin based off molecular weight and protein presence? Which samples measured from your ion exchange experiment had the most protein detected? Calculate the approximate pI of your proteins using the ion exchange Table 2. Ion Exchange Samples Tested by Bradford Assay Concentration BSA (μg/mL) Concentration BSA (mg/mL) Volume of Sample added to Bradford Reagent (μL) Absorbance at 595 nm Standard 1 250 0.25 50 0.32 Standard 2 125 0.125 50 0.16 Standard 3 62.5 0.0625 50 0.08 Standard 4 31.25 0.03125 50 0.04 Standard 5 15.625 0.015625 50 0.02 Blank 0.00 0.00 0.00 Table 3. Gel Filtration Fractions Tested by Bradford Assay Fraction #:…arrow_forward
- You are determining the concentration of your culture, and have counted the following on your hemocytometer as shown in Figure below: (note, blue cells have taken up Trypan blue & thus are dead. Red x's are cells that should not be counted because touching one of 2 sides of square). If your cells were diluted 1:2 before counting, what is the concentration of cells in your original culture?arrow_forwardWhat is meant by the statement “orthogonal analysis methods should be used for characterization of impurities”? Why do bacterial and plant cells require partial destruction prior to osmotic lysis? How can the partial destruction be achieved?arrow_forwardGradient PAGE uses differing gel concentrations along the length of the gel to achieve optimalseparation of proteins of a wide range of molecular weight proteins. How does the staining pattern of these types of gels differ from nongradient PAGE?arrow_forward
- In the extraction of DNA from a Banana, alcohol is added to the filtrate after filtration. What will happen if you do not filter the solution inside the zip lock prior to addition of cold alcohol?arrow_forwardWhich of the following correctly matches the chromatography technique with the molecular property being exploited for separation? ion-exchange: charge,affinity: polarity, gel-filtration: molecular mass ion-exchange: charge, affinity:molecular binding preference,gel-filtration: molecular size ion-exchange: charge,affinity: molecular binding preference,gel-filtration: isoilectric point ion-exchange: ion binding preference,affinity: molecular size,gel-filtration:polarityarrow_forwardThe diagrams below show the results of three FRAP experiments. Match the proteins to the statements. Protein B 1.0 0.8- 0.6- EVL 0.4- 0.2- 50 100 150 time (seconds) 0.2- 50 Protein A 100 150 time (seconds) 200 250 Protein that is completely tethered/anchored and does not move in the memebrane Protein that has some of it moving slowly while others are tethered/anchored in the membrane Protein that is fully mobile in the membrane Represent proteins associated with the cell membrane Represent cytoplasmic proteins that are moving around [Choose ] [Choose ] A, B and C None of these A Both A and B B с [Choose ] [Choose ] [Choose ] 200 250 50 Protein C 100 150 time (seconds) T 200 250arrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman
BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman
Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY
Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON
Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON

Biochemistry
Biochemistry
ISBN:9781319114671
Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.
Publisher:W. H. Freeman

Lehninger Principles of Biochemistry
Biochemistry
ISBN:9781464126116
Author:David L. Nelson, Michael M. Cox
Publisher:W. H. Freeman

Fundamentals of Biochemistry: Life at the Molecul...
Biochemistry
ISBN:9781118918401
Author:Donald Voet, Judith G. Voet, Charlotte W. Pratt
Publisher:WILEY

Biochemistry
Biochemistry
ISBN:9781305961135
Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougal
Publisher:Cengage Learning

Biochemistry
Biochemistry
ISBN:9781305577206
Author:Reginald H. Garrett, Charles M. Grisham
Publisher:Cengage Learning

Fundamentals of General, Organic, and Biological ...
Biochemistry
ISBN:9780134015187
Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. Peterson
Publisher:PEARSON