
Human Anatomy & Physiology (11th Edition)
11th Edition
ISBN: 9780134580999
Author: Elaine N. Marieb, Katja N. Hoehn
Publisher: PEARSON
expand_more
expand_more
format_list_bulleted
Concept explainers
Question
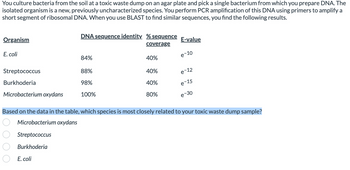
Transcribed Image Text:You culture bacteria from the soil at a toxic waste dump on an agar plate and pick a single bacterium from which you prepare DNA. The
isolated organism is a new, previously uncharacterized species. You perform PCR amplification of this DNA using primers to amplify a
short segment of ribosomal DNA. When you use BLAST to find similar sequences, you find the following results.
Organism
E. coli
Streptococcus
Burkhoderia
Microbacterium oxydans
DNA sequence identity. % sequence E-value
coverage
e-10
E. coli
84%
88%
98%
100%
40%
40%
40%
80%
e-12
e-15
e-30
Based on the data in the table, which species is most closely related to your toxic waste dump sample?
Microbacterium oxydans
Streptococcus
Burkhoderia
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
This is a popular solution
Trending nowThis is a popular solution!
Step by stepSolved in 3 steps

Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biology and related others by exploring similar questions and additional content below.Similar questions
- 1) Which technique is best suited to determining which genes are activated in a bacterium during infection while causing disease in a person. a) SDS-page b) microarray analysis c) RFLP analysis d) clone library analysis 2)Which of the following is not an application of PCR? a) Determine if two people are related. b) Identify a bacterial pathogen in a patient sample. c) Determine the gene sequence of the gene that codes for a bacterial enterotoxin. d) These are all applications of PCR.arrow_forwardWhy is taq polymerase used in pcr reactions.arrow_forwardIn Cohen-Boyer’s recombinant DNA procedure, ___i___ must be used for both the bacterial DNA and the amphibian DNA ___ii___ a) the same restriction enzyme; so that the restriction sites are identical in the DNA of each species b) different restriction enzymes; So that the genes outside the restriction site are maintained c) different restriction enzymes; to ensure that the newly introduced genes are maintained in the bacterial DNA d) the same restriction enzyme; to ensure that the newly formed DNA can replicatearrow_forward
- PCR primers are designed to only replicate the N gene sequence of the viral genome. Part of the N sequence we want to amplify is shown below. Typically, you design two primers, one to bind to each strand of the dsDNA. Copies are made from each strand so you get twice as much DNA from the PCR process. Potential primer locations are noted by the nucleotide sequences shown below. The PCR needs to make copies of the nucleotides shown in the middle (“87 nucleotides”). Remember the direction that DNA polymerase synthesizes new DNA strands. Select the two locations for the primers to bind and then fill in the correct sequence below the DNA sequence shown. You should have selected one location on each strand. Indicate the direction that the DNA polymerase (Taq polymerase) will move after binding to the primers in the attached imagearrow_forwardWhat are the reasons why there needs to be more than 10 cycles in the PCR process? Name 2 reasons.arrow_forwardBacteria and other microbes can be used to "clean up" an oil spill by breaking down oil into carbon dioxide and water. Two samples isolated from the Deepwater Horizon leak in the Gulf of Mexico were labeled A and B. The DNA of each was isolated and the percent thymine measured in each sample. Sample A contains 18.1% thymine and sample B contains 28.9% thymine. Assume the organisms contain normal double-stranded DNA and predict the composition of the other bases. percent adenine in sample A: SA percent guanine in sumple A: percent cytosine in sample A: percent adenine in sample B}: percent guanine in sample B 5 percent cytosine in sample B Beth samples are then denatured to remove the secondary structure. Which will have the higher temperature to densture!arrow_forward
- A DNA sequence is shown below, which includes a gene as marked. You have the restriction enzymes SalI and HindIII available to you to excise the gene prior to its incorporation into a plasmid vector. Which would you use to excise the gene?arrow_forwardWhat is a reason that a cut digest insert would not match up in length with a PCR insert in gel electrophoresis?arrow_forwardIn PCR, the size of the product is based on A) the position of the primers. B) the amount of DNA you add to the reaction. (c) the number of cycles the process has gone through. D) the amount of enzyme you add to the reaction. the location of the machine.arrow_forward
- After you design the PCR primers, you run the PCR on fluid from the patient’s nasal swab. Next, you need to evaluate the results. PCR makes billions of copies of just one sequence in a sample. Since you know the sequence, you also know the length (number of nucleotides) of the region you copied. There are about 130 nucleotides in the N gene fragment. This is typically stated as 130 base pairs. How can you visualize DNA and estimate its size? Load the DNA sample into an agarose gel (similar in consistency to jello) and apply an electric current to the gel. The DNA is charged and will move through the gel. The longer the DNA fragment, the more slowly it moves. The DNA is visualized by adding a fluorescent dye to your sample that sticks to DNA. When you look at the gel under UV light, the DNA should glow. What is the charge of a DNA molecule? Based on #1, would you expect DNA to be drawn to the (+) or (-) electrode of the gel electrophoresis chamber?arrow_forwardCan you help me with this question, please? What are the advantages of qPCR (RT-PCR) compared to conventional PCR? Choose all that apply a. human error is reduced as there are fewer human interactions with the samples b. you can visualize the results as the process is running c. samples can be compared as to the amount of template DNA in the original sample d. more samples can be run in a day by one personarrow_forwardA student is trying to add 9.0 pmol of primer mix to a 20.0 µL PCR. The primer mix is at a concentration of 50.0 µM, and the student determines that a serial dilution is required because directly adding the primer mix would require a volume less than 1.00 µL. The student takes 1.40 µL of the primer mix stock and mixes it with 25.0 µL of ultrapure H2O. Calculate the volume (in µL) of the intermediate solution required to add the correct amount of primer mix to the PCR.arrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 Human Anatomy & Physiology (11th Edition)BiologyISBN:9780134580999Author:Elaine N. Marieb, Katja N. HoehnPublisher:PEARSON
Human Anatomy & Physiology (11th Edition)BiologyISBN:9780134580999Author:Elaine N. Marieb, Katja N. HoehnPublisher:PEARSON Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax
Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax Anatomy & PhysiologyBiologyISBN:9781259398629Author:McKinley, Michael P., O'loughlin, Valerie Dean, Bidle, Theresa StouterPublisher:Mcgraw Hill Education,
Anatomy & PhysiologyBiologyISBN:9781259398629Author:McKinley, Michael P., O'loughlin, Valerie Dean, Bidle, Theresa StouterPublisher:Mcgraw Hill Education, Molecular Biology of the Cell (Sixth Edition)BiologyISBN:9780815344322Author:Bruce Alberts, Alexander D. Johnson, Julian Lewis, David Morgan, Martin Raff, Keith Roberts, Peter WalterPublisher:W. W. Norton & Company
Molecular Biology of the Cell (Sixth Edition)BiologyISBN:9780815344322Author:Bruce Alberts, Alexander D. Johnson, Julian Lewis, David Morgan, Martin Raff, Keith Roberts, Peter WalterPublisher:W. W. Norton & Company Laboratory Manual For Human Anatomy & PhysiologyBiologyISBN:9781260159363Author:Martin, Terry R., Prentice-craver, CynthiaPublisher:McGraw-Hill Publishing Co.
Laboratory Manual For Human Anatomy & PhysiologyBiologyISBN:9781260159363Author:Martin, Terry R., Prentice-craver, CynthiaPublisher:McGraw-Hill Publishing Co. Inquiry Into Life (16th Edition)BiologyISBN:9781260231700Author:Sylvia S. Mader, Michael WindelspechtPublisher:McGraw Hill Education
Inquiry Into Life (16th Edition)BiologyISBN:9781260231700Author:Sylvia S. Mader, Michael WindelspechtPublisher:McGraw Hill Education

Human Anatomy & Physiology (11th Edition)
Biology
ISBN:9780134580999
Author:Elaine N. Marieb, Katja N. Hoehn
Publisher:PEARSON

Biology 2e
Biology
ISBN:9781947172517
Author:Matthew Douglas, Jung Choi, Mary Ann Clark
Publisher:OpenStax

Anatomy & Physiology
Biology
ISBN:9781259398629
Author:McKinley, Michael P., O'loughlin, Valerie Dean, Bidle, Theresa Stouter
Publisher:Mcgraw Hill Education,

Molecular Biology of the Cell (Sixth Edition)
Biology
ISBN:9780815344322
Author:Bruce Alberts, Alexander D. Johnson, Julian Lewis, David Morgan, Martin Raff, Keith Roberts, Peter Walter
Publisher:W. W. Norton & Company

Laboratory Manual For Human Anatomy & Physiology
Biology
ISBN:9781260159363
Author:Martin, Terry R., Prentice-craver, Cynthia
Publisher:McGraw-Hill Publishing Co.

Inquiry Into Life (16th Edition)
Biology
ISBN:9781260231700
Author:Sylvia S. Mader, Michael Windelspecht
Publisher:McGraw Hill Education