
Human Anatomy & Physiology (11th Edition)
11th Edition
ISBN: 9780134580999
Author: Elaine N. Marieb, Katja N. Hoehn
Publisher: PEARSON
expand_more
expand_more
format_list_bulleted
Concept explainers
Question
Transcribed Image Text:## Step 1
The branch of Biological Sciences that deals with the study of genomes is known as Genomics. The genome is the complete set of genetic information present in an organism.
As per the guidelines, the answers for the first 3 questions are being provided here. The student is requested to upload the remaining questions separately.
## Step 2
### Question 1:
GWAS or Genome-wide Association Study is a method for identifying the genes responsible for giving an organism its phenotype. This method is very useful because:
- It provides information on the SNPs that need attention to understand the genetic risk for a condition in an organism.
- SNP identification helps to understand the mechanisms causing genetic risk, providing clarity on differences between alleles.
- This method can find genomic variants that cause a particular trait or disease in an individual.
GWAS can solve problems such as:
- Finding out genes associated with a particular complex disease.
- Screening a large number of SNPs from the genome of an individual.
- Understanding the genetic mechanisms of disease better.
### Question 2:
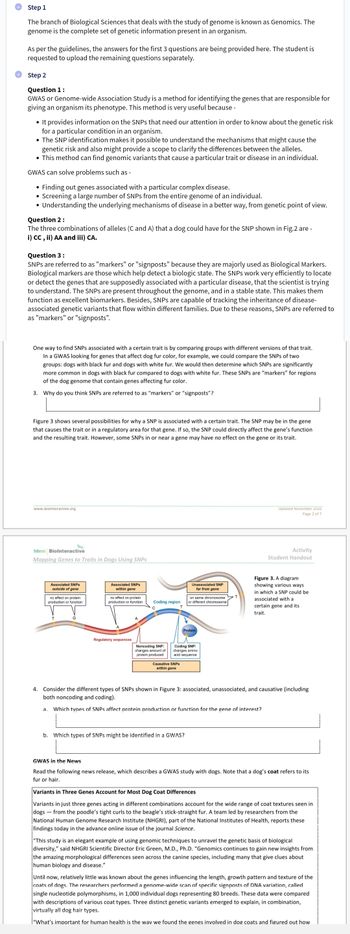
The combinations of alleles (C and A) that a dog could have for the SNP shown in Fig. 2 are:
i) CC
ii) AA
iii) CA
### Question 3:
SNPs are referred to as "markers" or "signposts" because they are majorly used as Biological Markers that help detect a biological state. SNPs efficiently locate or detect genes associated with particular diseases. Present throughout the genome, SNPs act as excellent biomarkers and can track the inheritance of disease-associated genetic variants within families.
One way to find SNPs associated with a trait is by comparing groups with different versions of that trait. For example, in dogs, comparing black coats to white coats reveals informative SNPs associated with color. These SNPs are "markers" for regions containing genes affecting fur color.
3. Why do you think SNPs are referred to as “markers” or “signposts”?
### Diagram Analysis (Figure 3)
**Figure 3 Description:**
- The diagram shows various ways an SNP could associate with a gene and its trait.
- It illustrates SNPs:
- **Associated & Outside of Gene:** No effect on protein function.
- **Associated & Within Gene:** No effect or it may change protein production/function.
- **Unassociated & Far from Gene or Within Gene:** No effect.
- **Causative SNPs Within Gene:** Affect
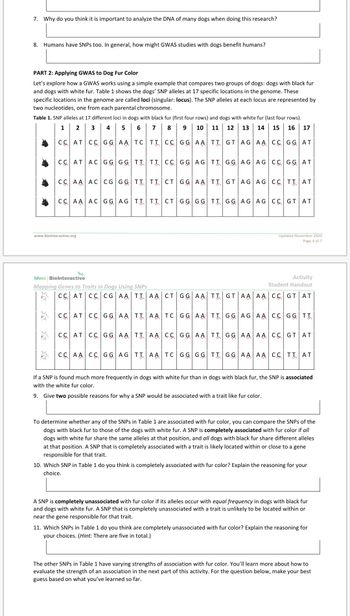
Transcribed Image Text:**PART 2: Applying GWAS to Dog Fur Color**
Let's explore how a GWAS (Genome-Wide Association Study) works using a simple example that compares two groups of dogs: dogs with black fur and dogs with white fur. Table 1 shows the dogs’ SNP (Single Nucleotide Polymorphism) alleles at 17 specific locations in the genome. These specific locations in the genome are called loci (singular: locus). The SNP alleles at each locus are represented by two nucleotides, one from each parental chromosome.
**Table 1.** SNP alleles at 17 different loci in dogs with black fur (first four rows) and dogs with white fur (last four rows).
| | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 |
|---|----|----|----|----|----|----|----|----|----|----|----|----|----|----|----|----|----|
|
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
This is a popular solution
Trending nowThis is a popular solution!
Step by stepSolved in 4 steps

Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biology and related others by exploring similar questions and additional content below.Similar questions
- The beach mouse (Peromyscus polionotus) is a small rodent found in the southeastern United States. Coat color in the beach mouse is variable, from dark brown to blonde (Figure 1). A pigment called eumelanin is responsible for some of the diversity in mouse coat color. The MC1R gene, located on chromosome 8, plays a major role in the synthesis of eumelanin. There are two alleles of MC1R: R allele. Results in a functional MC1R protein that leads to stimulation of pigment production. This allele is found in dark colored mice. C allele. Results in a non-functional MC1R protein. Pigment production is, as a result, not stimulated. This allele is found in light colored mice. Figure 1. Beach mouse with light fur (left) and dark (right). These mice are hunted by visual predators like owls and fox. The distribution of coat color is not random. Beach mice with dark fur are found further inland, on dark brown soils while mice with light fur are found on beaches, where the sand is light brown.…arrow_forwardThe majority of GWAS associated variants exist in non-coding regions. This has led to additional challenges in understanding the biological mechanisms behind the trait, as the associated variants may not have a clear impact on gene function. Explain two non-coding mechanisms and how they contribute to genetic variation. In your answer, mention what types of sequencing data would assist in determining whether a non-coding GWAS locus may operate under these mechanisms.arrow_forwardThe beach mouse (Peromyscus polionotus) is a small rodent found in the southeastern United States. Coat color in the beach mouse is variable, from dark brown to blonde (Figure 1). A pigment called eumelanin is responsible for some of the diversity in mouse coat color. The MC1R gene, located on chromosome 8, plays a major role in the synthesis of eumelanin. There are two alleles of MC1R: R allele. Results in a functional MC1R protein that leads to stimulation of pigment production. This allele is found in dark colored mice. C allele. Results in a non-functional MC1R protein. Pigment production is, as a result, not stimulated. This allele is found in light colored mice. Figure 1. Beach mouse with light fur (left) and dark (right). These mice are hunted by visual predators like owls and fox. The distribution of coat color is not random. Beach mice with dark fur are found further inland, on dark brown soils while mice with light fur are found on beaches, where the sand is…arrow_forward
- 24. 23andme is a direct to consumer genotyping company that uses a microarray to genotype people on 1 million known SNPS in the genome. A man who was adopted wants to know about his ancestry, and purchases a 23andme kit for himself. He finds that of each autosome pair, exactly one is classified as 100% Sub-Saharan African, while the other is a mixture of various East Asian and European components. He also finds that his X chromosome is 100% Sub-Saharan African. What can this man confidently infer about his recent ancestry? a. His mother was of Sub-Saharan African descent b. His father was of Sub-Saharan African descent c. He has no Neanderthal ancestry d. He has no mutations relative to his parents e. 23andme mixed up two samples and he can't infer anythingarrow_forwardBlue eye color allele (b) is recessive to the dominant to the black eye color allele (B). When we sequence around the locus of the blue gene from 10 pure breed species and 10 black pure breed species we notice difference in dna sequence. The black eye species have short tandem repeat (STR), which is a short DNA sequence that is repeated 6 times in the species with blue eyes, and 10 times in the species with black eyes. These repeats are only 4 cM apart from the orange gene locus. You design a PC test that distinguishes between the 6 and 10 STRs by electrophoresis. Make pedigree that will let us to identify which alleles of the blue gene was the dominant and or recessive?arrow_forwardDNA markers have greatly enhanced the mapping of genes in humans. What are DNA markers, and what advantage do they confer?arrow_forward
- Why does linkage disequilibrium represent an advantage and a disadvantage for mapping genetic variants associated with complex traits?arrow_forwardIn the early 1970s, Igor Dawid and Antonie Blacklerconducted classical experiments that first showed directly the maternal inheritance of mtDNA in vertebrates.Their studies used crosses between two closely relatedspecies of frogs, Xenopus laevis and Xenopus borealis,which have mtDNAs that vary in many nucleotides. Thetechniques they used at the time were not sensitiveenough to detect small amounts of paternal DNA. Whattechniques that are highly sensitive to small amounts ofDNA could be used today? How could you use thesetechniques to determine if paternal mitochondrial DNAwas present in the progeny of the interspecies cross?arrow_forwardYou are studying celiacs disease and have identified a potential gene for the disease using linkage analysis. You decide to test your hypothesis by making transgenic mice that carry a nonsense mutation in the candidate gene you have identified to see if they develop a phenotype similar to that found in Celiacs disease What sort or experimental approach are you using when you test your gene in your mouse model? a. Reverse genetic b. Quantitative genetic C. Forward genetic D.Expression analysis E. None of the abovearrow_forward
- 1) Using the diagram below, sketch in the pattern of bands you would expect to see after digesting the DNA of the TT, Tt and tt genotypes of the TAS2R38 gene. Use the Base Pair Standards on the left of the diagram as a reference in drawing the positions of the bands. Base Pair TASTER GENO TYPE Standards (Base Pairs) TT Tt tt 500 400 300 200 100 50arrow_forwardIn your opinion, is antibiotic use at farm sites promote the development and spread of antibiotic resistant genes in bacteria? Why or why not please explain based on the articlearrow_forwardIn the Human Genome Project, researchers have collectedlinkage data from many crosses in which the male washeterozygous for molecular markers and many crosses wherethe female was heterozygous for the markers. The distancebetween the same two markers, computed in map units, isdifferent between males and females. In other words, thelinkage maps for human males and females are not the same.Propose an explanation for this discrepancy. Do you think thesizes of chromosomes (excluding the Y chromosome) in humanmales and females are different? How could physical mappingresolve this discrepancy?arrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 Human Anatomy & Physiology (11th Edition)BiologyISBN:9780134580999Author:Elaine N. Marieb, Katja N. HoehnPublisher:PEARSON
Human Anatomy & Physiology (11th Edition)BiologyISBN:9780134580999Author:Elaine N. Marieb, Katja N. HoehnPublisher:PEARSON Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax
Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax Anatomy & PhysiologyBiologyISBN:9781259398629Author:McKinley, Michael P., O'loughlin, Valerie Dean, Bidle, Theresa StouterPublisher:Mcgraw Hill Education,
Anatomy & PhysiologyBiologyISBN:9781259398629Author:McKinley, Michael P., O'loughlin, Valerie Dean, Bidle, Theresa StouterPublisher:Mcgraw Hill Education, Molecular Biology of the Cell (Sixth Edition)BiologyISBN:9780815344322Author:Bruce Alberts, Alexander D. Johnson, Julian Lewis, David Morgan, Martin Raff, Keith Roberts, Peter WalterPublisher:W. W. Norton & Company
Molecular Biology of the Cell (Sixth Edition)BiologyISBN:9780815344322Author:Bruce Alberts, Alexander D. Johnson, Julian Lewis, David Morgan, Martin Raff, Keith Roberts, Peter WalterPublisher:W. W. Norton & Company Laboratory Manual For Human Anatomy & PhysiologyBiologyISBN:9781260159363Author:Martin, Terry R., Prentice-craver, CynthiaPublisher:McGraw-Hill Publishing Co.
Laboratory Manual For Human Anatomy & PhysiologyBiologyISBN:9781260159363Author:Martin, Terry R., Prentice-craver, CynthiaPublisher:McGraw-Hill Publishing Co. Inquiry Into Life (16th Edition)BiologyISBN:9781260231700Author:Sylvia S. Mader, Michael WindelspechtPublisher:McGraw Hill Education
Inquiry Into Life (16th Edition)BiologyISBN:9781260231700Author:Sylvia S. Mader, Michael WindelspechtPublisher:McGraw Hill Education

Human Anatomy & Physiology (11th Edition)
Biology
ISBN:9780134580999
Author:Elaine N. Marieb, Katja N. Hoehn
Publisher:PEARSON

Biology 2e
Biology
ISBN:9781947172517
Author:Matthew Douglas, Jung Choi, Mary Ann Clark
Publisher:OpenStax

Anatomy & Physiology
Biology
ISBN:9781259398629
Author:McKinley, Michael P., O'loughlin, Valerie Dean, Bidle, Theresa Stouter
Publisher:Mcgraw Hill Education,

Molecular Biology of the Cell (Sixth Edition)
Biology
ISBN:9780815344322
Author:Bruce Alberts, Alexander D. Johnson, Julian Lewis, David Morgan, Martin Raff, Keith Roberts, Peter Walter
Publisher:W. W. Norton & Company

Laboratory Manual For Human Anatomy & Physiology
Biology
ISBN:9781260159363
Author:Martin, Terry R., Prentice-craver, Cynthia
Publisher:McGraw-Hill Publishing Co.

Inquiry Into Life (16th Edition)
Biology
ISBN:9781260231700
Author:Sylvia S. Mader, Michael Windelspecht
Publisher:McGraw Hill Education