
Biochemistry
9th Edition
ISBN: 9781319114671
Author: Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.
Publisher: W. H. Freeman
expand_more
expand_more
format_list_bulleted
Question
Suppose that there is a protein consisting of two polypeptide chains with the given sequences in the picture.
Before performing Edman sequencing method, it is required to identify the protein's amino acid composition first. Explain why it is required to do the said step first before doing Edman sequencing method.
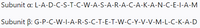
Transcribed Image Text:Subunit a: L-A-D-C-S-T-C-W-A-S-A-R-A-C-A-K-A-N-C-E-I-A-M
Subunit B: G-P-C-W-I-A-R-S-C-T-E-T-W-C-Y-V-V-M-L-C-K-A-D
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
Step by stepSolved in 3 steps

Knowledge Booster
Similar questions
- Explain how the use of radiolabeled amino acids in this procedure helped to reveal the genetic code.arrow_forwardAlign two sequences: horizontal – GGAATGG, vertical – ATG, m=1, mm = 0, g=-1. Use the table below for the NW matrix. Write down and score all optimal global alignments. Complete the NW matrix below and show the alignment paths. Use the arrows and circles for the matrix and paths. Click the shapes and move them using your keyboard arrow keys (or drag). Click the shapes and then right-click to copy/paste or click and use Ctrl + C to copy, then Ctrl + V to paste (in Windows MS Word). 0000 Align and score 4 optimal alignments here. the first line, v sequence in the second line and Each alignment should have h sequence individual scores for each alignment position in the third line.arrow_forwardTotal nucleic acids are extracted from a growing culture of yeast cells. They are then mixed with specialized beads to which the single-stranded DNA molecule with sequence 5’-TTTTTTTTTTTTTTTTTTTTTTTTT-3’ has been covalently attached to the surface (see image to the right, where each black line represents a polynucleotide sequence). After a short incubation time, the beads are removed from the mixture. When you analyze the cellular nucleic acids stuck to the beads, which type of nucleic acid (i.e. DNA, rRNA, etc.) do you expect to be the most abundant? Why?arrow_forward
- You have been given an unknown low weight polypeptide to sequence. Luckily, you have a plentiful supply. You are given a limited amount of Edmond's reagent-just enough to perform 1 restricted digestion for every fragment that you generate but not for complete digestion of every fragment. You have the following enzymes and reagents available to do your digestion analysis: Chymotrypsin, V8, HCI (1M), Cyanogen Bromide (CnBr) and 50mM of ammonium bicarbonate. You are only given enough Carboxypeptidase for one fragment analysis. You fortunately have a plentiful supply of ion chromatography columns, eluting buffers, and SDS-PAGE gels to help you separate individual amino acids for analysis and fragment size (length) determination. And you have a really good spec and an HPLC. From the information given below, you will need to construct the sequence of your peptide! You also have the following information: 1. Digestion of the intact polypeptide with 1M HCl will liberate all amino acids (you…arrow_forwardGive typing answer with explanation and conclusion to all parts Maxim-Gilbert and Sanger Sequencing are two different methods used to sequence DNA. Describe the general techniques of Maxim-Gilbert and Sanger DNA Sequencing. List the advantages and disadvantages of each.arrow_forwardFrom the results of your BLAST search you can link to the GENE entry for one of your top hits. This link is located under the “Related Information” heading at the right hand side of each displayed alignment (i.e. scroll down to the“Alignments” section). Q1: How many exons and introns are annotated for this gene? Q2: What is the function of the encoded protein?arrow_forward
- You wish to align a 300 bp nucleotide sequence to a genome. Which of the following pairwise sequence alignment algorithms is BEST suited for this task? Global alignment or Local alignment? Please explain or elaborate why you chose this alignment algorithm.arrow_forwardExplain why a higher agarose concentration is preferred when seperating smaller DNA molecules. Include mathematical analysis in your answer.arrow_forwardhow would you identify whether or not complete resolution of the three amino acids has been achievedarrow_forward
- When the cDNA was sequenced by the Sanger method utilizing ddCTP, the following products were obtained: Tetranucleotide Hexanucleotide Nonanucleotide Decanucleotide Dodenucleotide Octadecanucleotide Nonadecanucleotide 21-nucleotide 6c. What is the sequence of the bases in the mRNA coding for the peptide above? Thearrow_forwardPlease explain how to use the global alignment algorithm: Needleman-Wunsch. Use the match 3, mismatch -3, linear gap penalty -3. Protein sequence 1: PHWG Protein sequence 2: HWEGarrow_forwardTranscriptome analysis involves two separate methodologies: gene expression and RNA seq analyses. The 10 items below are a scrambled listing of the steps used in the two procedures. Identify the steps involved in RNA seq from the list below. Use the numbers in the list to refer to each step. Once the steps for RNA seq have been identified, write the steps in the order in which they are performed during the experiment. (1) DNA sequencing (2) Allow for hybridization and wash excess cRNA. (3) Mix labeled cRNA with array chip. (4) PCR amplification (5) Measure fluorescence intensity to determine abundance of transcripts. (6) Add labeled cRNA at each microarray location. (7) Map cDNA sequences to the genome of the organism to determine identity and abundance of transcripts. (8) mRNA isolation from cells (9) Prepare fluorescently labeled cRNA probes (10) cDNA synthesisarrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman
BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman
Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY
Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON
Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON

Biochemistry
Biochemistry
ISBN:9781319114671
Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.
Publisher:W. H. Freeman

Lehninger Principles of Biochemistry
Biochemistry
ISBN:9781464126116
Author:David L. Nelson, Michael M. Cox
Publisher:W. H. Freeman

Fundamentals of Biochemistry: Life at the Molecul...
Biochemistry
ISBN:9781118918401
Author:Donald Voet, Judith G. Voet, Charlotte W. Pratt
Publisher:WILEY

Biochemistry
Biochemistry
ISBN:9781305961135
Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougal
Publisher:Cengage Learning

Biochemistry
Biochemistry
ISBN:9781305577206
Author:Reginald H. Garrett, Charles M. Grisham
Publisher:Cengage Learning

Fundamentals of General, Organic, and Biological ...
Biochemistry
ISBN:9780134015187
Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. Peterson
Publisher:PEARSON