
Biochemistry
9th Edition
ISBN: 9781319114671
Author: Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.
Publisher: W. H. Freeman
expand_more
expand_more
format_list_bulleted
Concept explainers
Question
thumb_up100%
Transcribed Image Text:66%
12:42 AM
Nucleic Acids.p...
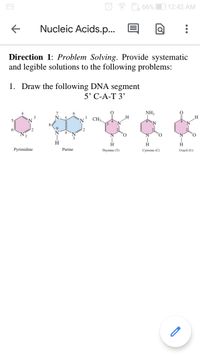
Direction 1: Problem Solving. Provide systematic
and legible solutions to the following problems:
1. Draw the following DNA segment
5' С-А-Т 3'
NH2
CH3
5
N
12
H
H
H
Pyrimidine
Purine
Thymine (T)
Cytosine (C)
Uracil (U)
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
Step by stepSolved in 2 steps with 1 images

Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biochemistry and related others by exploring similar questions and additional content below.Similar questions
- E Threonine 6. You have identified some intermediates in threonine synthesis: A, B, C, D and E. You grow a few of your mutants in the presence of these different intermediates to determine the order in which the gene products act. Below are your results. A (+) means growth and a (-) means no growth. Given these data, draw the best possible pathway for the synthesis of threonine. The diagram should use arrows to indicate one intermediate being changed to another intermediate. Indicate which gene produces the product responsible for the conversion by listing the mutant in that gene above the arrow. Mt1 Mt2 Mt4 Mt7arrow_forwarda. Why is it important to eat food containing antioxidants? Write at leasttwo reactions to prove the answer. b. Write in four ways the following nucleotide sequence: ATGCA. c. Explain i. Hoogsteen pairing & ii. Hyperchromic effectiii. Epimers Iv. Mutarotaton v. Aldose vi. Anomers vii. Mutarotationarrow_forwardI need the answer as soon as possiblearrow_forward
- 1. complete the table below: Volume Procedure (mL) Crude 120 extract GFC 20 IEX 10 Protein Total Unit Total Specific content protein Activity Activity Activity (mg/mL) (mg) (U/mL) (U) (U/mg) 20 (a) (d) 0.356 (h) 11 (b) (e) 0.207 (i) 9 (c) (f) (g) (j) % Purification Recovery fold (k) (1) (n) (m) (0)arrow_forwardThe vast majority of structures deposited in the Protein Databank (>95%) have been determined using NMR Hard keratins (such as in the skin) are less resistant to stretch because the larger number of disulfide bonds resist deformative forces. Structures produced by X-ray diffraction are as good as the solutions from which they are derived they are false and I know that but explain why they are falsearrow_forward6. b. Draw a box around the disulfide bridge in oxytocin, if present, or write "none". 7. Mark each peptide bond in oxytocin by making the corresponding line in the structure thicker or marking it with a different color. The first one is shown for you as an example (in dark orange). 8. Number the central carbon of each amino acid in oxytocin by pointing a small arrow to it or by circling the corresponding vertex in the image. Numbers 1 and 2 indicate the central carbons of the first and second amino acids of oxytocin, and are shown for you as an example. 9. Fill out the following table, listing amino acids that make up oxytocin in order, from the N terminus to the C terminus, characterizing each amino acid by the properties of its R group (side chains), and briefly indicating the reasoning for the characterization. You may consult amino acid groupings by category in the slides (or the textbook, p.49), but you must explain the reasoning for each in your own words. CO 1 AA# Abbre- Full…arrow_forward
- Penicillin G is a natural antibiotic that is useful for treating infections caused by Gram positive bacteria. What is the functional benefit of the semi-synthetic antimicrobials carbenicillin and ampicillin, generated by chemical modification of the R group so their R groups each are a bit different than the R group seen with penicillin G?arrow_forwardonly need zarrow_forwardWhich one of the following statements about nucleic acids is correct? Multiple Choice NTPs can be incorporated into either DNA or RNA molecules, but dNTPs can only be incorporated into DNA molecules A nitrogenous base, 5-carbon sugar, and two phosphate groups would most accurately be referred to as a nucleotide diphosphate. During nucleic acid synthesis, the phosphate group at the 3' carbon of the incoming monomer is covalently linked to the hydroxyl group at the 5' carbon of the existing macromolecule. RNA and DNA have all the same pyrimidine bases in them, but differ in the selection of purine bases. A nucleic acid found in a cell with the sequence GCCTTATCGCG would most likely be found paired with another molecule in double-stranded form.arrow_forward
- 5. A mixture containing phenylalanine, tryptophan, and leucine was subjected to electrophoresis. Determine which of the amino acids moved the farthest distance assuming that the experiment was performed at the pH indicated: (a) pH 6.0 (b) pH 5.0arrow_forwardI'm very confused on #13. Can anyone explain if the claim is valid or not? I don't understand how to calculate #15. Can someone show the formula for it? #16 I think is C and #17 I think is B. Is this correct?arrow_forward1. You need 0.5 liter of a 0.1 M buffer for a biochemical reaction that you plan to perform at pH 2.0. On the shelf in your laboratory you have available phosphoric acid, and the amino acids aspartate, glycine, glutamate, tyrosine, and histidine (all you have available are the fully deprotonated forms of the molecules); these are the only buffer materials you have available. (The pKa values for these compounds are given below.) For phosphoric acid, pKal1 = 2.12, pKa2 = 7.21, and pKa3 = 12.32 For aspartate, pKaCOOH = 1.88, pKaNH3+ = 9.60, pKa sidechain = 3.65 For glycine, pKaCOOH = 2.34, pKaNH3+ = 9.60 For glutamate, pKaCOOH = 2.19, pKaNH3+ = 9.67, pKa sidechain = 4.25 For histidine, pKɑCOOH = 1.82, pKɑNH3+ = 9.17, pKa sidechain = 6.00 For tyrosine, pKaCOOH = 2.20, pKaNH3+ = 9.11, pKa sidechain = 10.07 a. Of these possible buffers, which is likely to yield the greatest resistance to pH change from the starting pH of 2.0, assuming that you were going to run a biochemical reaction that…arrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman
BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman
Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY
Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON
Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON

Biochemistry
Biochemistry
ISBN:9781319114671
Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.
Publisher:W. H. Freeman

Lehninger Principles of Biochemistry
Biochemistry
ISBN:9781464126116
Author:David L. Nelson, Michael M. Cox
Publisher:W. H. Freeman

Fundamentals of Biochemistry: Life at the Molecul...
Biochemistry
ISBN:9781118918401
Author:Donald Voet, Judith G. Voet, Charlotte W. Pratt
Publisher:WILEY

Biochemistry
Biochemistry
ISBN:9781305961135
Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougal
Publisher:Cengage Learning

Biochemistry
Biochemistry
ISBN:9781305577206
Author:Reginald H. Garrett, Charles M. Grisham
Publisher:Cengage Learning

Fundamentals of General, Organic, and Biological ...
Biochemistry
ISBN:9780134015187
Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. Peterson
Publisher:PEARSON