
Biochemistry
9th Edition
ISBN: 9781319114671
Author: Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.
Publisher: W. H. Freeman
expand_more
expand_more
format_list_bulleted
Question

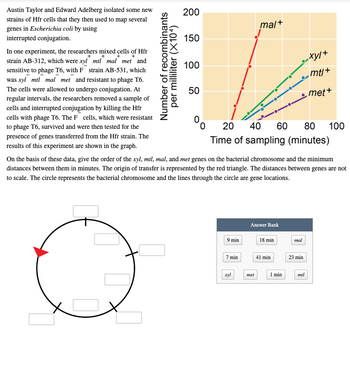
Transcribed Image Text:Austin Taylor and Edward Adelberg isolated some new
strains of Hfr cells that they then used to map several
200
mal+
genes in Escherichia coli by using
interrupted conjugation.
150
In one experiment, the researchers mixed cells of Hfr
strain AB-312, which were xyl* mtl* mal* met* and
sensitive to phage T6, with F strain AB-531, which
was xyl mtl mal met and resistant to phage T6. The
100
mt/+
cells were allowed to undergo conjugation. At regular
intervals, the researchers removed a sample of cells and
50
met+
interrupted conjugation by killing the Hfr cells with
phage T6. The F cells, which were resistant to phage
T6, survived and were then tested for the presence of
0.
20
40
60
80
100
genes transferred from the Hfr strain. The results of this
experiment are shown in the graph.
Time of sampling (minutes)
On the basis of these data, give the order of the xyl, mtl, mal, and met genes on the bacterial chromosome and the minimum
distances between them in minutes. The origin of transfer is represented by the red triangle. The distances between genes are not
to scale. The circle represents the bacterial chromosome and the lines through the circle are gene locations.
Number of recombinants
per milliliter (X104)
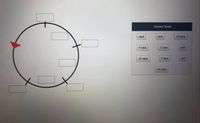
Transcribed Image Text:Answer Bank
mal
met
18 min
9 min
1 min
mtl
23 min
7 min
xyl
41 min
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
This is a popular solution
Trending nowThis is a popular solution!
Step by stepSolved in 2 steps with 2 images

Follow-up Questions
Read through expert solutions to related follow-up questions below.
Follow-up Question
The answer bank only has one 9 minute option, therefore 9 minutes can only be used once. Is it possible that there is a mistake in the above solution?
Transcribed Image Text:### Mapping Bacterial Genes Using Interrupted Conjugation
Austin Taylor and Edward Adelberg isolated new strains of Hfr (High frequency recombination) cells to map several genes in *Escherichia coli* by using interrupted conjugation.
In one experiment, the researchers mixed cells of Hfr strain AB-312, which were \( \text{xyl}^+ \text{mtl}^+ \text{mal}^+ \text{met}^+ \) and sensitive to phage T6, with \( F^- \) strain AB-531, which was \( \text{xyl}^- \text{mtl}^- \text{mal}^- \text{met}^- \) and resistant to phage T6. The cells were allowed to undergo conjugation. At regular intervals, the researchers removed a sample of cells and interrupted conjugation by killing the Hfr cells with phage T6. The \( F^- \) cells, which were resistant to phage T6, were then tested for the presence of genes transferred from the Hfr strain. The results of this experiment are shown in the graph.
### Experiment Results Graph
The graph plots the "Number of recombinants per milliliter (\( \times 10^4 \))" against "Time of sampling (minutes)”. The four genes’ appearance on the graph is tracked as follows:
- \( \text{mal}^+ \) starts appearing at around 20 minutes, increasing steadily over time.
- \( \text{xyl}^+ \) begins to show at around 40 minutes.
- \( \text{mtl}^+ \) appears at around 60 minutes.
- \( \text{met}^+ \) is the last to show up, starting around 80 minutes.
### Gene Order and Distances
Based on these data, the order of the genes on the bacterial chromosome and the minimum distances between them in minutes are derived. The origin of transfer is marked by a red triangle. The circle represents the bacterial chromosome, and the lines through the circle indicate gene locations.
The expected order of genes and their respective distances are:
- \( \text{mal} \) at 1 minute
- \( \text{m} (position to be specified using the given bank)
- \( \text{xyl} \)
- \( \text{mtl} \)
- \( \text{met
Solution
by Bartleby Expert
Follow-up Questions
Read through expert solutions to related follow-up questions below.
Follow-up Question
The answer bank only has one 9 minute option, therefore 9 minutes can only be used once. Is it possible that there is a mistake in the above solution?

Transcribed Image Text:### Mapping Bacterial Genes Using Interrupted Conjugation
Austin Taylor and Edward Adelberg isolated new strains of Hfr (High frequency recombination) cells to map several genes in *Escherichia coli* by using interrupted conjugation.
In one experiment, the researchers mixed cells of Hfr strain AB-312, which were \( \text{xyl}^+ \text{mtl}^+ \text{mal}^+ \text{met}^+ \) and sensitive to phage T6, with \( F^- \) strain AB-531, which was \( \text{xyl}^- \text{mtl}^- \text{mal}^- \text{met}^- \) and resistant to phage T6. The cells were allowed to undergo conjugation. At regular intervals, the researchers removed a sample of cells and interrupted conjugation by killing the Hfr cells with phage T6. The \( F^- \) cells, which were resistant to phage T6, were then tested for the presence of genes transferred from the Hfr strain. The results of this experiment are shown in the graph.
### Experiment Results Graph
The graph plots the "Number of recombinants per milliliter (\( \times 10^4 \))" against "Time of sampling (minutes)”. The four genes’ appearance on the graph is tracked as follows:
- \( \text{mal}^+ \) starts appearing at around 20 minutes, increasing steadily over time.
- \( \text{xyl}^+ \) begins to show at around 40 minutes.
- \( \text{mtl}^+ \) appears at around 60 minutes.
- \( \text{met}^+ \) is the last to show up, starting around 80 minutes.
### Gene Order and Distances
Based on these data, the order of the genes on the bacterial chromosome and the minimum distances between them in minutes are derived. The origin of transfer is marked by a red triangle. The circle represents the bacterial chromosome, and the lines through the circle indicate gene locations.
The expected order of genes and their respective distances are:
- \( \text{mal} \) at 1 minute
- \( \text{m} (position to be specified using the given bank)
- \( \text{xyl} \)
- \( \text{mtl} \)
- \( \text{met
Solution
by Bartleby Expert
Knowledge Booster
Similar questions
- order: 1 Gm Depakote On hand: 500 mg/tablet How many tablets will you administer for the ordered dose?arrow_forwardIdentify the following labeled structures.arrow_forwardDoctor ordered Haloperidol 3mg po bid. You have scored tablets containing Haloperidol 2mg. How many tab will you give?arrow_forward
- a 4YO 35lb child has an order for ceftazidime 40 mg/kg IV every 8 hours. How many mg will you administer per dose?arrow_forwardConsider the following two nonhomologous wildtype chromosomes, where letters or numbers represent genes, the "-" represents the centromere of each chromosome, and chromosomes are shown on separate lines. ABCDE-FGHIJK 123-45678 Identify the type of rearrangement shown in each of the following (A-C) and then identify whether it is balanced or unbalanced. Assume that the individual is diploid and heterozygous for the rearrangement. A. • ABCDE-FGHIJKGH 123-45678 Rearrangement: [Select] • Balanced or Unbalanced: [Select] B. • ABCDGF-EHIJK 123-45678 Rearrangement ✓ [Select] • Balanced or Un pericentric inversion Robertsonian translocation deletion tandem duplicate paracentric inversion C. reciprocal translocation ABCDE dispersed duplicate nonreciprocal translocation 123-45arrow_forward17 18 19 21 22 X Y 多、 00 20arrow_forward
- Beta-tubulin has a molecular mass of 55 kDa, Approximately how many amino acids are found in Beta-tubulin? O 45 O 450 O 4,500 O 45.000 O 450.000arrow_forwardA child has been prescribed pethidine 35mg IM at 0800 hours. 1ml Stock ampoules have been labelled 5% pethidine. What volume must be drawn up for injection? ?????? ????????= ???????ℎ ????????/????? ???????ℎ x (volume of stock solution)arrow_forwardA series of bovine serum albumin (BSA) was prepared and 1 mL of each solution was subjected to a Bradford assay. The increase in absorbance at 595 nm relative to the concentration of protein was plotted as shown below. Save 1.0- 0.8- 0.6- 0.4- 0.2- 0.04 0.0 0.5 1.0 2.0 2.5 1.5 BSA concentration (mg/mL) a) Using the graph, calculate the original concentration of protein present in a mixture of haemoglobin and methylene blue (diluted 1:50), which gave an absorbance of 0.20 at a wavelength of 595 nm using the Bradford assay. Show all workings. Absorbance (O.D.)arrow_forward
- 1-2 tsp po qid #8 oz How many days will the medication last?arrow_forward9201703 aitavia bns standzus omne ledet avode margsib ad: nf is Lari valab ni aidaxo babivong magsih art griau nont (d al gastos to sengug il blow weig 100 etantadija-emysne stb sew to rotse olup warb vies (5 563 is isbom ysx 8. Define, give an example of, and explain both Catabolism and Anabolism.arrow_forwardLM 1200 LM200) LM (1200 Identify the formed element labeled "a." %3Darrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman
BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman
Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY
Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON
Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON

Biochemistry
Biochemistry
ISBN:9781319114671
Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.
Publisher:W. H. Freeman

Lehninger Principles of Biochemistry
Biochemistry
ISBN:9781464126116
Author:David L. Nelson, Michael M. Cox
Publisher:W. H. Freeman

Fundamentals of Biochemistry: Life at the Molecul...
Biochemistry
ISBN:9781118918401
Author:Donald Voet, Judith G. Voet, Charlotte W. Pratt
Publisher:WILEY

Biochemistry
Biochemistry
ISBN:9781305961135
Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougal
Publisher:Cengage Learning

Biochemistry
Biochemistry
ISBN:9781305577206
Author:Reginald H. Garrett, Charles M. Grisham
Publisher:Cengage Learning

Fundamentals of General, Organic, and Biological ...
Biochemistry
ISBN:9780134015187
Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. Peterson
Publisher:PEARSON