
Biology Today and Tomorrow without Physiology (MindTap Course List)
5th Edition
ISBN: 9781305117396
Author: Cecie Starr, Christine Evers, Lisa Starr
Publisher: Cengage Learning
expand_more
expand_more
format_list_bulleted
Question
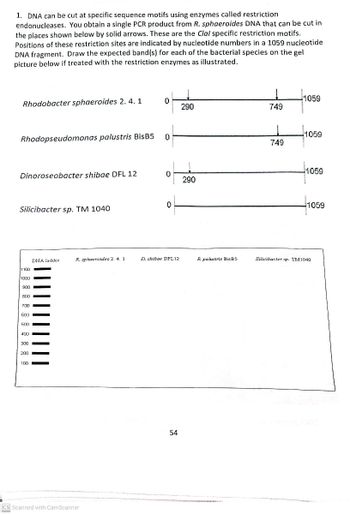
Transcribed Image Text:1. DNA can be cut at specific sequence motifs using enzymes called restriction
endonucleases. You obtain a single PCR product from R. sphaeroides DNA that can be cut in
the places shown below by solid arrows. These are the Clal specific restriction motifs.
Positions of these restriction sites are indicated by nucleotide numbers in a 1059 nucleotide
DNA fragment. Draw the expected band(s) for each of the bacterial species on the gel
picture below if treated with the restriction enzymes as illustrated.
Rhodobacter sphaeroides 2. 4.1
1059
0
290
749
Rhodopseudomonas palustris BisB5
0
Dinoroseobacter shibae DFL 12
0
290
Silicibacter sp. TM 1040
0
1059
749
1059
1059
DNA ladder
R. sphaeroides 2. 4. 1
D. shibae DFL12
R. palustris BisB5
Silicibacter sp. TM1040
1100
1000
900
800
700
600
500
400
300
200
100
||||||||
CS Scanned with CamScanner
54
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
Step by stepSolved in 2 steps with 1 images

Knowledge Booster
Similar questions
- You learned in the chapter that an STR locus is a locus where alleles differ in the number of copies of a short, tandemly repeated DNA sequence. PCR is used to determine the number of alleles present, as shown by the size of the DNA fragment amplified. In the Figure below are the results of PCR analysis for STR alleles at a locus where the repeat unit length is 9 bp, and alleles are known that have 5 to 11 copies of the repeat. Given the STR alleles present in the adults, state whether each of the four juveniles could or could not be an off-spring of those two adults. Explain your answers.arrow_forwardTranscribe and translate the following DNA sequence (nontemplate strand): 5’-ATGGCCGGTTATTAAGCA-3’arrow_forwardWhy are antibiotic resistance markers such as ampR important components of bacterial plasmid cloning vectors? a. The plasmid must have resistance to accept DNA inserts. b. They allow the detection of plasmids that contain an inserted DNA fragment. c. They ensure the presence of the ori site. d. They ensure that the plasmid can be cut by a restriction enzyme. e. They allow identification of bacteria that have taken up a plasmid.arrow_forward
- Match the method with the appropriate enzyme. _____ PCR a. Taq polymerase _____ cutting DNA b. DNA ligase _____ cDNA synthesis c. reverse transcriptase _____ DNA sequencing d. restriction enzyme _____ pasting DNA e. DNA polymerase (not Taq)arrow_forwardChemical Mutagenesis of DNA Bases Show the nucleotide sequence changes that might arise in a dsDNA (coding strand segment GCTA) upon mutagenesis with (a) HNO2, (b) bromouracil, and (C) 2-aminopurine.arrow_forwardWhat Art the Features of the Series of -omes? Define the following terms: a. Genome b. Transcriptome c. Proteome d. Metabolome e. Fluxomearrow_forward
- Analyzing Cloned Sequences A base change (A to T) is the mutational event that created the mutant sickle cell anemia allele of beta globin. This mutation destroys an MstII restriction site normally present in the beta globin gene. This difference between the normal allele and the mutant allele can be detected with Southern blotting. Using a labeled beta globin gene as a probe, what differences would you expect to see for a Southern blot of the normal beta globin gene and the mutant sickle cell gene?arrow_forwardn gel electrophoresis of DNA, the different bands in the final gel form because the DNA molecules _______. a. are from different organisms b. have different lengths c. have different nucleotide compositions d. have different genesarrow_forwardAn individuals set of unique _______ can be used as a DNA profile. a. DNA sequence c. short tandem repeats b. SNPs d. all of the abovearrow_forward
- An individuals set of unique _______ can be used in DNA profiling. a. DNA sequences b. short tandem repeats c. SNPs d. all of the abovearrow_forwardWhen Griffith injected mice with a combination of live rough-strain and heat-killed smooth-strain pneumococci, he discovered that (a) the mice were unharmed (b) the dead mice contained living rough-strain bacteria (c) the dead mice contained living smooth-strain bacteria (d) DNA had been transferred from the smooth-strain bacteria to the mice (e) DNA had been transferred from the rough-strain bacteria to the smooth-strain bacteriaarrow_forwardDuring proofreading, which of the following enzymes reads the DNA? primase topoisomerase DNA pol helicasearrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning Biology: The Unity and Diversity of Life (MindTap...BiologyISBN:9781305073951Author:Cecie Starr, Ralph Taggart, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology: The Unity and Diversity of Life (MindTap...BiologyISBN:9781305073951Author:Cecie Starr, Ralph Taggart, Christine Evers, Lisa StarrPublisher:Cengage Learning Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning
Biology: The Dynamic Science (MindTap Course List)BiologyISBN:9781305389892Author:Peter J. Russell, Paul E. Hertz, Beverly McMillanPublisher:Cengage Learning Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College
Concepts of BiologyBiologyISBN:9781938168116Author:Samantha Fowler, Rebecca Roush, James WisePublisher:OpenStax College Biology: The Unity and Diversity of Life (MindTap...BiologyISBN:9781337408332Author:Cecie Starr, Ralph Taggart, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology: The Unity and Diversity of Life (MindTap...BiologyISBN:9781337408332Author:Cecie Starr, Ralph Taggart, Christine Evers, Lisa StarrPublisher:Cengage Learning

Biology Today and Tomorrow without Physiology (Mi...
Biology
ISBN:9781305117396
Author:Cecie Starr, Christine Evers, Lisa Starr
Publisher:Cengage Learning

Biology: The Unity and Diversity of Life (MindTap...
Biology
ISBN:9781305073951
Author:Cecie Starr, Ralph Taggart, Christine Evers, Lisa Starr
Publisher:Cengage Learning

Biology: The Dynamic Science (MindTap Course List)
Biology
ISBN:9781305389892
Author:Peter J. Russell, Paul E. Hertz, Beverly McMillan
Publisher:Cengage Learning

Human Heredity: Principles and Issues (MindTap Co...
Biology
ISBN:9781305251052
Author:Michael Cummings
Publisher:Cengage Learning

Concepts of Biology
Biology
ISBN:9781938168116
Author:Samantha Fowler, Rebecca Roush, James Wise
Publisher:OpenStax College

Biology: The Unity and Diversity of Life (MindTap...
Biology
ISBN:9781337408332
Author:Cecie Starr, Ralph Taggart, Christine Evers, Lisa Starr
Publisher:Cengage Learning