
Case Studies In Health Information Management
3rd Edition
ISBN: 9781337676908
Author: SCHNERING
Publisher: Cengage
expand_more
expand_more
format_list_bulleted
Question
thumb_up100%
Genetics Q8
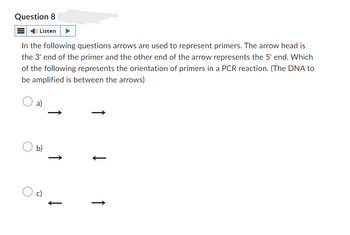
Transcribed Image Text:Question 8
Listen
In the following questions arrows are used to represent primers. The arrow head is
the 3' end of the primer and the other end of the arrow represents the 5' end. Which
of the following represents the orientation of primers in a PCR reaction. (The DNA to
be amplified is between the arrows)
a)
b)
↑
c)
↑
↑
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
Step by stepSolved in 2 steps with 1 images

Knowledge Booster
Similar questions
- Quinolone antibiotics treat bacterial infections by blocking the activity of topoisomerase. Why does this treatment work? Explain what occurs at the molecular level.arrow_forwardWhat are Okazaki fragments and how they are formed?arrow_forwardGenetics in Practice case studies are critical-thinking exercises that allow you to apply your new knowledge of human genetics to real-life problems. Case study Michelle was a 42-year-old woman who had declined counselling and amniocentesis at 16 weeks of pregnancy but was referred for genetic counseling after an abnormal ultrasound at 20 weeks of gestation. After the ultrasound, a number of findings suggested a possible chromosome abnormality in the fetus. The ultrasound showed swelling under the skin at the back of the fetuss neck; shortness of the femur, humerus, and ear length; and underdevelopment of the middle section of the fifth finger. Michelles physician performed an amniocentesis and referred her to the genetics program. Michelle and her husband did not want genetic counseling before receiving the results of the cytogenetic analysis. This was Michelles third pregnancy; she and her husband, Mike, had a 6-year-old daughter and a 3-year-old son. At their next session, the counselor informed the couple that the results revealed trisomy 21, explored their understanding of Down syndrome, and elicited their experiences with people with disabilities. She also reviewed the clinical concerns revealed by the ultrasound and associated anomalies (mild to severe intellectual disability, cardiac defects, and kidney problems). The options available to the couple were outlined. They were provided with a booklet written for parents making choices after the prenatal diagnosis of Down syndrome. After a week of careful deliberation with their family, friends, and clergy, they elected to terminate the pregnancy. Should physicians discourage a 42-year-old woman from having children because of an increased chance of a chromosomal abnormality?arrow_forward
- Provide a brief summary of the Sanger sequencing method.arrow_forwardAnswers to all problems are at the end of this book. Detailed solutions are available in the Student Solutions Manual, Study Guide, and Problems Book. B- and Z-DNA Helical Parameters I A 41.5-nm-long duplex DNA molecule in the B conformation adopts the A-conformation upon dehydration. How long is it now? What is its approximate number of base pairs?arrow_forwardYou are in charge of a new gene therapy clinic. Two cases have been referred to you for review and possible therapy. Case 1. A mutation in the promoter of a proto-oncogene causes the gene to make too much of its normal product, a receptor protein that promotes cell division. The uncontrolled cell division has caused cancer. Case 2. A mutation in an exon of a tumor-suppressor gene makes this gene nonfunctional. The product of this gene normally suppresses cell division. The mutant gene cannot suppress cell division, and this has led to cancer. What treatment options can you suggest for each case?arrow_forward
- Why were radioactive sulfur and phosphorous used to label bacteriophage in Hershey and Chase’s experiments?arrow_forwardWhich of the following does the enzyme primase synthesize? DNA primer RNA primer Okazaki fragments phosphodiester linkagearrow_forwardDNA Profiles as Tools for Identification A PCR-based paternity test is conducted using STRs that consistently produce a unique DNA fragment pattern from a single chromosome. Examining the results of the following Southern blot, which male(s) can be excluded as the father of the child? Which male(s) could be the father of the child?arrow_forward
- Which of the following best describes the process of DNA sequencing? a. DNA is separated on a gel, and the different bands are labeled with fluorescent nucleotides and scanned with a laser. b. A laser is used to fluorescently label the nucleotides present within the DNA, the DNA is run on a gel, and then the DNA is broken into fragments. c. Nucleotides are scanned with a laser and incorporated into the DNA that has been separated on a gel, and then the DNA is amplified with PCR. d. Fragments of DNA are produced in a reaction that labels them with any of four different fluorescent dyes, and the fragments then are run on a gel and scanned with a laser. e. DNA is broken down into its constituent nucleotides, and the nucleotides are then run on a gel and purified with a laser.arrow_forwardAnswers to all problems are at the end of this book. Detailed solutions are available in the Student Solutions Manual, Study Guide, and Problems Book. DNA Supercoiling Parameters A “relaxed,� circular, double-stranded DNA molecule (1600 bp) is in a solution where conditions favor 10 bp per turn. What is the value of L0 for this DNA molecule? Suppose DNA gyrase introduces 12 negative supercoils into this molecule. What are the values of L, W, and T now? What is the superhelical density, ?arrow_forwardA beginning genetics student is attempting to complete an assignment to draw a base pair from a DNA molecule. The drawing is incomplete, and the student does not know how to finish. He asks for your advice. The assignment sheet shows that the drawing is to contain three hydrogen bonds, a purine, and a pyrimidine. From your knowledge of the pairing rules and the number of hydrogen bonds in A/T and G/C base pairs, what base pair do you help the student draw?arrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
- Case Studies In Health Information ManagementBiologyISBN:9781337676908Author:SCHNERINGPublisher:Cengage
 Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning
Biology Today and Tomorrow without Physiology (Mi...BiologyISBN:9781305117396Author:Cecie Starr, Christine Evers, Lisa StarrPublisher:Cengage Learning  Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax
Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage LearningEssentials of Pharmacology for Health ProfessionsNursingISBN:9781305441620Author:WOODROWPublisher:Cengage
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage LearningEssentials of Pharmacology for Health ProfessionsNursingISBN:9781305441620Author:WOODROWPublisher:Cengage

Case Studies In Health Information Management
Biology
ISBN:9781337676908
Author:SCHNERING
Publisher:Cengage

Biology Today and Tomorrow without Physiology (Mi...
Biology
ISBN:9781305117396
Author:Cecie Starr, Christine Evers, Lisa Starr
Publisher:Cengage Learning


Biology 2e
Biology
ISBN:9781947172517
Author:Matthew Douglas, Jung Choi, Mary Ann Clark
Publisher:OpenStax

Human Heredity: Principles and Issues (MindTap Co...
Biology
ISBN:9781305251052
Author:Michael Cummings
Publisher:Cengage Learning

Essentials of Pharmacology for Health Professions
Nursing
ISBN:9781305441620
Author:WOODROW
Publisher:Cengage