
Human Heredity: Principles and Issues (MindTap Course List)
11th Edition
ISBN: 9781305251052
Author: Michael Cummings
Publisher: Cengage Learning
expand_more
expand_more
format_list_bulleted
Concept explainers
Question
Need help, think is C
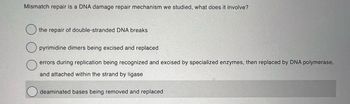
Transcribed Image Text:Mismatch repair is a DNA damage repair mechanism we studied, what does it involve?
the repair of double-stranded DNA breaks
pyrimidine dimers being excised and replaced
errors during replication being recognized and excised by specialized enzymes, then replaced by DNA polymerase,
and attached within the strand by ligase
deaminated bases being removed and replaced
Expert Solution
arrow_forward
Step 1
DNA Repair:
DNA repair is defined as the system of cellular responses which are associated with the restoration of the normal base-pair sequence and structure of damaged DNA.
Types of DNA Repair mechanisms:
The types of DNA repair mechanisms include base excision repair, nucleotide excision repair, mismatch repair, homologous recombination and non-homologous end joining.
Step by stepSolved in 2 steps

Knowledge Booster
Learn more about
Need a deep-dive on the concept behind this application? Look no further. Learn more about this topic, biology and related others by exploring similar questions and additional content below.Similar questions
- What is an advantage and a disadvantage of this repair system?arrow_forwardDuring DNA replication, the function of RNA primers is to Group of answer choices serve as a binding site for DNA ligase separate the two strands of the double helix to open replication "bubbles" serve as starting points for DNA strand elongation by DNA polymerase in the 3' - 5' direction prevent new-separated strands of DNA from rejoining serve as starting points for DNA strand elongation in the 5' - 3' direction by DNA polymerasearrow_forwardDuring DNA replication, one of the new strands of DNA is synthesized continuously, while the other is synthesized as a number of separate fragments of DNA that are subsequently linked by DNA ligase. This is because O replication starts at many points on the chromosome RNA primers only anneal to one of the parental strands of DNA one of the parental strands is unwound slower than the other by helicase DNA polymerase III only synthesizes DNA in the 5' - 3' directionarrow_forward
- I’m having trouble which is not a correct statement. Can you pose explain this problem?arrow_forwardDNA polymerases are processive, which means that they remain tightly associated with the template strand while moving rapidly and adding nucleotides to the growing daughter stand. Which piece of the replication machinery accounts for this characteristic? Helicase Sliding Clamp Single Stranded Binding Protein Primasearrow_forwardMatch the enzymes involved in DNA replication with their function. Primase [ Choose ] [ Choose] Synthesizes short RNA segment to initiate new DNA strand Helicase Main enzyme that extends RNA primer by adding DNA nucleotides to it Stabilizes single-stranded DNA Relieves over-winding of DNA ahead of the replication fork Removes RNA primers preceding Okazaki fragment and replaces RNA nucleotides with DNA nucleotides Single-stranded binding proteins Unwinds DNA helix Synthesizes the ends of the linear chromosome Seals nicks between adjacent DNA segments DNA polymerase III [ Choose ] DNA polymerasel [ Choose ] DNA Ligase [ Choose ] Topoisomerase [ Choose ]arrow_forward
- In DNA replication, the fundamental reason that Okasaki fragments are produced is that Group of answer choices DNA replication can proceed only in the 5’ to 3’ direction replication on the leading strand is slower than replication on the lagging strand DNA polymerase tends to slip while replicating the lagging strand DNA polymerase has to alternate between the leading and lagging strands during replication none of these are true.arrow_forwardsomething is wrong with replisome shown in this cartoon -what do you think it is? Can you spot the problem?arrow_forwardWhich statement is true about dna replicationarrow_forward
- The proteins and enzymes listed below are all required for DNA replication in E. coli, but they are listed in a random order. Determine the correct order in which they function in replication, by selecting the correct number from the drop-down menu in each case, with 1 being first and 6 being last.arrow_forwardWhat is a replication fork?arrow_forwardMatch the bold DNA repair response(s) to the triggering type of DNA damage. Homologous Recombination Mishmatch Repair Base Excision Repair Nucleotide Excision Repair Non-homologous End Joining Single-strand DNA breaks Removal of repair lesions such as photoproducts caused by UV including Thymine dimers Double-Strand Break repair mechanism which is an accurate repair mechanism without any introduction of insertions or deletions. It requires a sister chromatid as a template. This repair mechanism uses just DNA glycosylase to remove Uracil (no other enzymes or complexes are required) then DNA polymerase can use the template stand to add the complementary base where the Uracil has been removedInterstrand Crosslink Repair This repair mechanism is used to recognize and repair mis-incorporation of base that can arise during DNA replication. Removal and replacement of modifying bases such as Uracil, 8-hyroxyguanine and others. Double-strand Break that is termed as “Quick and Dirty” as it is…arrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning
Human Heredity: Principles and Issues (MindTap Co...BiologyISBN:9781305251052Author:Michael CummingsPublisher:Cengage Learning

Human Heredity: Principles and Issues (MindTap Co...
Biology
ISBN:9781305251052
Author:Michael Cummings
Publisher:Cengage Learning