
Concept explainers
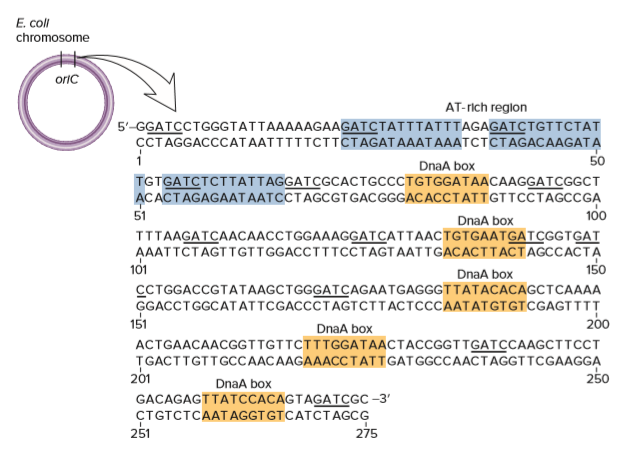
As shown, five DnaA boxes are found within the origin of replication in E. coli. Take a look at these five sequences carefully.
A. Are the sequences of the five DnaA boxes very similar to each other? (Hint: Remember that DNA is double-stranded; think about these sequences in the forward and reverse directions.)
B. What is the most common sequence for a DnaA box? In other words, what is the most common base in the first position, second position, and so on until the ninth position? The most common sequence is called the consensus sequence.
C. The E. coli chromosome is about 4.6 million bp long. Based on random chance, is it likely that the consensus sequence for a DnaA box occurs elsewhere in the E. coli chromosome? If so, why aren’t there multiple origins of replication in E. coli?
Trending nowThis is a popular solution!
Step by stepSolved in 2 steps

- For linear 34,000 kb DNA calculate a) contour length; b) the length of DNA as packaged in nucleosomes and c) the length of the DNA in a 30-nm fiber.arrow_forwardThe A and G compositions (mole percent) of one of the strands of a duplex DNA is A = 27 and G = 30. (a) What would be the T and C compositions of the complementary strand? (b) What can be said about the A and G compositions of its complementary strand?arrow_forwarda. what is the nucleotide sequence of the DNA chain synthesized from the primer. label the 5' to 3' ends b. what is the nucleotide sequence of the DNA chain use as the template strand. label the 5' and 3' ends c. write out the nucleotide sequence of the DA double helix (label the 5' and 3' ends)arrow_forward
- In the drawing shown below, the top strand (purple) is the template DNA, and the bottom shows the synthesis of the lagging strand (DNA in blue and RNA primers in red). 3' 5" ыш Left Okazaki fragment --- -11-- Middle Okazaki fragment Right Okazaki fragment 5' 3' After DNA polymerase I removes the middle primer and fills in with DNA, where is DNA ligase needed? See the arrows on both sides of the middle primer. Explain whether ligase is needed at the left arrow, at the right arrow, or both?arrow_forwardWhat part(s) of a nucleotide (namely, phosphate, sugar, and/orbase) is(are) found in the major and minor grooves of doublestrandedDNA, and what part(s) is(are) found in the DNA backbone?If a DNA-binding protein does not recognize a specificnucleotide sequence, do you expect that it binds to the majorgroove, the minor groove, or the DNA backbone? Explain.arrow_forwardCan you please help with 1c please picture with 1 graph is for question 1a) picture with 4 graphs is for question 1b) 1a) E. coli DNA and binturong DNA are both 50% G-C. If you randomly shear E. coli DNA into 1000 bp fragments and put it through density gradient equilibrium centrifugation, you will find that all the DNA bands at the same place in the gradient, and if you graph the distribution of DNA fragments in the gradient you will get a single peak (see below). If you perform the same experiment with binturong DNA, you will find that a small fraction of the DNA fragments band separately in the gradient (at a different density) and give rise to a small "satellite" peak on a graph of the distribution of DNA fragments in the gradient (see below). Why do these two DNA samples give different results, when they're both 50% G-C? 1b) If you denatured the random 1000 bp fragments of binturong DNA that you produced in question 1a by heating them to 95ºC, and then cooled them down to 60ºC…arrow_forward
- Even though the eukaryotic genome is thousands of times larger than the prokaryotic genome, DNA replication times are relatively similar. Explain how this is possible.arrow_forwardUsing the figure below, what is molecule "A" (type a 1, 2 or 3 in the blank) nuclease ligase DNA polymerase What is the function of molecule "A"? to separate the double helix into two to piece together the Okazaki segments to copy the new DNA strand to the old strand by complementary base pairing Using the figure below, what is molecule "G" (type a 1, 2 or 3 in the blank) nuclease ligase DNA polymerase What is the function of molecule "G"? to separate the double helix into two to piecearrow_forwardHow would I calculate sanger sequencing fragments that will produce greenarrow_forward
- All ingredients required for the synthesis of DNA were placed in a test tube. The primer has the sequence 5’-TCGATCA-3’. First determine where the primer will bind to the DNA template given below (Do this by writing the sequence of the primer on top of the template – make sure you find a location that is complementary in sequence and that the primer is antiparallel with respect to the template) and then indicate the nucleotides that DNA polymerase would attach onto the primer as it synthesizes DNA (hint: in which direction does DNA polymerase attach nucleotides onto a primer?) See figure 13.16 in book. 5’-GACGTAGTCTGATGCTAGCATGCTGATCGAAAGAG-3’arrow_forwardWhich of the following pairs of base sequences could form ashort stretch of a normal double helix of DNA?(A) 5′-AGCT-3′ with 5′-TCGA-3′(B) 5′-GCGC-3′ with 5′-TATA-3′(C) 5′-ATGC-3′ with 5′-GCAT-3′(D) All of these pairs are correctarrow_forwardCan you please help with 1f please picture with 1 graph is for question 1a) picture with 4 graphs is for question 1b) 1a) E. coli DNA and binturong DNA are both 50% G-C. If you randomly shear E. coli DNA into 1000 bp fragments and put it through density gradient equilibrium centrifugation, you will find that all the DNA bands at the same place in the gradient, and if you graph the distribution of DNA fragments in the gradient you will get a single peak (see below). If you perform the same experiment with binturong DNA, you will find that a small fraction of the DNA fragments band separately in the gradient (at a different density) and give rise to a small "satellite" peak on a graph of the distribution of DNA fragments in the gradient (see below). Why do these two DNA samples give different results, when they're both 50% G-C? 1b) If you denatured the random 1000 bp fragments of binturong DNA that you produced in question 1a by heating them to 95ºC, and then cooled them down to 60ºC and…arrow_forward
 Human Anatomy & Physiology (11th Edition)BiologyISBN:9780134580999Author:Elaine N. Marieb, Katja N. HoehnPublisher:PEARSON
Human Anatomy & Physiology (11th Edition)BiologyISBN:9780134580999Author:Elaine N. Marieb, Katja N. HoehnPublisher:PEARSON Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax
Biology 2eBiologyISBN:9781947172517Author:Matthew Douglas, Jung Choi, Mary Ann ClarkPublisher:OpenStax Anatomy & PhysiologyBiologyISBN:9781259398629Author:McKinley, Michael P., O'loughlin, Valerie Dean, Bidle, Theresa StouterPublisher:Mcgraw Hill Education,
Anatomy & PhysiologyBiologyISBN:9781259398629Author:McKinley, Michael P., O'loughlin, Valerie Dean, Bidle, Theresa StouterPublisher:Mcgraw Hill Education, Molecular Biology of the Cell (Sixth Edition)BiologyISBN:9780815344322Author:Bruce Alberts, Alexander D. Johnson, Julian Lewis, David Morgan, Martin Raff, Keith Roberts, Peter WalterPublisher:W. W. Norton & Company
Molecular Biology of the Cell (Sixth Edition)BiologyISBN:9780815344322Author:Bruce Alberts, Alexander D. Johnson, Julian Lewis, David Morgan, Martin Raff, Keith Roberts, Peter WalterPublisher:W. W. Norton & Company Laboratory Manual For Human Anatomy & PhysiologyBiologyISBN:9781260159363Author:Martin, Terry R., Prentice-craver, CynthiaPublisher:McGraw-Hill Publishing Co.
Laboratory Manual For Human Anatomy & PhysiologyBiologyISBN:9781260159363Author:Martin, Terry R., Prentice-craver, CynthiaPublisher:McGraw-Hill Publishing Co. Inquiry Into Life (16th Edition)BiologyISBN:9781260231700Author:Sylvia S. Mader, Michael WindelspechtPublisher:McGraw Hill Education
Inquiry Into Life (16th Edition)BiologyISBN:9781260231700Author:Sylvia S. Mader, Michael WindelspechtPublisher:McGraw Hill Education





