
Biochemistry
9th Edition
ISBN: 9781319114671
Author: Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.
Publisher: W. H. Freeman
expand_more
expand_more
format_list_bulleted
Question
thumb_up100%
Below is kinetic data obtained for an enzyme-catalyzed reaction. The enzyme concentration is fixed at 100 nM.
Using a Lineweaver-Burke plot, calculate the catalytic efficiency for this reaction. Report your answer in scientific notation to three significant figures in units of M-1s-1.
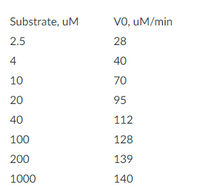
Transcribed Image Text:Substrate, uM
VO, uM/min
2.5
28
4
40
10
70
20
95
40
112
100
128
200
139
1000
140
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
This is a popular solution
Trending nowThis is a popular solution!
Step by stepSolved in 5 steps with 1 images

Knowledge Booster
Similar questions
- The following question focuses on how the parameters regulating enzyme function might change, and how these might appear graphically on a Michaelis-Menten plot and a Lineweaver-Burke plot. Carbonic anhydrase is an enzyme that will convert CO2 and water into HCO3. CO2 + H20 > H+ + HCO3 There are many different isoforms of this enzyme. (see for instance http://en.wikipedia.org/wiki/Carbonic_anhydrase . Imidazol is a competitive inhibitor of carbonic anhydrase. It is effective at an alkaline (high) pH; in lower (more acidic) pH, it no longer inhibits the enzyme. Draw on a separate graph a Lineweaver-Burke plot for the effects of this compound at high pH and low pH. Be sure to label the axes and put in sample data points.arrow_forwardPlease answer A & B attached with image.arrow_forwardThe following question focuses on how the parameters regulating enzyme function might change, and how these might appear graphically on a Michaelis-Menten plot and a Lineweaver-Burke plot. Carbonic anhydrase is an enzyme that will convert CO2 and water into HCO3. CO2 + H20 > H+ + HCO3 There are many different isoforms of this enzyme. (see for instance http://en.wikipedia.org/wiki/Carbonic_anhydrase . Assume that one variant has a Km of 10 µM and a different variant has a Km of 100 µM. Draw on the same graph a typical Michaelis-Menton plot showing the alteration in the rate of carbonic anhydrase as the CO2 level is varied for the two different variants of enzyme, assuming the concentration of the enzyme (10 mM) in the test tube is kept constant. Assume that you have equal amounts of the two different variants of carbonic anhydrase in a number of test tubes and that the Vmax for both enzymes are the same. Be sure to label the axes. For the same conditions as above, draw a…arrow_forward
- Create a ONE page, formal summary, including APA Formatted in-text citations, of the enzyme amalyze and how it is used in the meat processing industry (Address all guiding questions below in your summary). Note, references can be the second page of the assignment. Include the questions in ur answer Which enzyme is used in the industry (if more than one enzyme is included, you can choose one of the enzymes) Describe the molecular structure of the enzyme (how many amino acids; discuss different levels of protein structure-secondary, tertiary, quaternary, key structural components/amino acids in active site) Describe how the enzyme works in the industry and why it is a key component for this industry. What specific reaction(s) are being catalyzed? What types of bonds are being broken?arrow_forwardA particular reaction has a ΔG‡ of 30.0 kJ mol-1 at 25.0 °C. In the presence of an enzyme, the same reaction has a ΔG‡ of 1.50 kJ mol-1 at the same temperature. Calculate the rate enhancement of this enzyme. (R = 8.3145 J mol-1 K-1)arrow_forwardPlease handraw this graph with all the necessary detailed information: Imagine that I text enzyme rate for four different temperatures: 10 degrees celsius, 20 degrees celsisus, 30 degree celsius, and 40 degree celsius, in separate tubes. The enzyme appears to work faster as temperature increases, but completely ceases activity at 40 degrees celcius. Sketch a graph to show this outcome, but here you will graph product formation (nmoles/mL) vs. time (minutes). The graph should be 4 lines and HANDDRAWN. Include a legend if necessary. You do not need precise quantitivate values, but most show the correct trends on the graph.arrow_forward
- Using the appropriate graph and table above, explain what the R48C mutation appears to be doing to the enzyme’s function. Discuss the kinetic parameter changes and their meaning in this context, not the structure of the enzyme, which was not given to you.arrow_forwardPlease don't provide handwritten solution ....arrow_forwardAn uncatalyzed reaction has keq=50. in the presence of an appropriate enzyme.the forward rate of the reaction increased by 20-fold.what is the equilibrium constant in the presence of the enzyme?arrow_forward
- The following data was obtained during kinetic analysis of an enzyme with and without an inhibitor. Substrate concentration (mM) Reaction rate without inhibitor (µM/s) Reaction rate with inhibitor (µM/s) 10 28 12 20 50 23 40 83 42 60 107 58 100 139 83 200 179 125 300 197 150 400 209 167 560 227 197 How do you calculate the KM for the enzyme in the absence of an inhibitor. And how do you calculate kcat with the given enzymatic concentration of 5 µM.arrow_forwardBelow is kinetic data obtained for an enzyme-catalyzed reaction. The enzyme concentration is fixed at 100 nM. Using a Lineweaver-Burke plot, calculate the kcat value for this reaction. Report your answer to three significant figures in units of 1/sec.arrow_forwardThe rate constants of an enzyme-catalyzed reaction, obeying the Michaelis-Menten kinetics, have been determined : E + S K₁ = 2 x 108 M-¹ S-¹ -1 -1 -1 K-₁= 1 x 10³ S K₂ = 5 x 10³ S-1 K₁ 1 K-1 ES K₂ E +P 1- Determine the Michaelis constant Km of the enzyme. 2- Determine the catalytic constant (kcat) of the enzyme. 3- Determine the catalytic efficiency of the enzyme.arrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman
BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman
Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY
Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON
Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON

Biochemistry
Biochemistry
ISBN:9781319114671
Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.
Publisher:W. H. Freeman

Lehninger Principles of Biochemistry
Biochemistry
ISBN:9781464126116
Author:David L. Nelson, Michael M. Cox
Publisher:W. H. Freeman

Fundamentals of Biochemistry: Life at the Molecul...
Biochemistry
ISBN:9781118918401
Author:Donald Voet, Judith G. Voet, Charlotte W. Pratt
Publisher:WILEY

Biochemistry
Biochemistry
ISBN:9781305961135
Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougal
Publisher:Cengage Learning

Biochemistry
Biochemistry
ISBN:9781305577206
Author:Reginald H. Garrett, Charles M. Grisham
Publisher:Cengage Learning

Fundamentals of General, Organic, and Biological ...
Biochemistry
ISBN:9780134015187
Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. Peterson
Publisher:PEARSON