
Biochemistry
9th Edition
ISBN: 9781319114671
Author: Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.
Publisher: W. H. Freeman
expand_more
expand_more
format_list_bulleted
Question
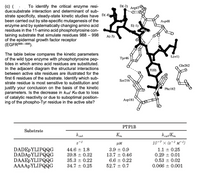
Transcribed Image Text:(c) (
due:substrate interaction and determinant of sub-
To identify the critical enzyme resi-
D(-2)
Arg47
strate specificity, steady-state kinetic studies have D(-4).
been carried out by site-specific mutagenesis of the
enzyme and by systematically changing amino acid
residues in the 11-amino acid phosphotyrosine con-
taining substrate that simulate residues 988 – 998
of the epidermal growth factor receptor
(EGFR988–998).
Asp48
E(-1)
A(-3)
The table below compares the kinetic parameters
of the wild type enzyme with phosphotyrosine pep-
tides in which amino acid residues are substituted.
In the adjacent diagram the structural interactions
L(+1)
Tyr46
Gln262
Y(0)
between active site residues are illustrated for the
first 6 residues of the substrate. Identify which sub-
strate residue is most sensitive to substitution and
Ser216
justify your conclusion on the basis of the kinetic
parameters. Is the decrease in kcat/ KM due to loss
of catalytic reactivity or due to suboptimal position-
ing of the phospho-Tyr residue in the active site?
Phe182
Asp181
PTP1B
Substrate
kcat
K.
kcat/Km
JUM
10-7 x (s-1 M1)
DADEPYLIPQQG
DADAPYLIPQQG
DAAEPYLIPQQG
AAAAPYLIPQQG
44.6 ± 1.8
39.8 + 0.32
3.9 + 0.9
13.7 + 0.46
1.1 + 0.25
0.29 + 0.01
35.3 + 0.22
6.6 ± 0.22
0.53 + 0.02
34.7 + 0.25
52.7 + 0.7
0.066 0.001
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
Step by stepSolved in 4 steps with 9 images

Knowledge Booster
Similar questions
- asap plsarrow_forward1CFD CALCIUM-FREE CALMODULIN How long is the protein’s primary sequence?Does this protein have any secondary, tertiary and/or quaternary structures present?Are there any molecular additions to this protein that are not amino acarrow_forwardQuestion in Image, thanks!arrow_forward
arrow_back_ios
arrow_forward_ios
Recommended textbooks for you
 BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman
BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman
Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY
Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON
Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON

Biochemistry
Biochemistry
ISBN:9781319114671
Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.
Publisher:W. H. Freeman

Lehninger Principles of Biochemistry
Biochemistry
ISBN:9781464126116
Author:David L. Nelson, Michael M. Cox
Publisher:W. H. Freeman

Fundamentals of Biochemistry: Life at the Molecul...
Biochemistry
ISBN:9781118918401
Author:Donald Voet, Judith G. Voet, Charlotte W. Pratt
Publisher:WILEY

Biochemistry
Biochemistry
ISBN:9781305961135
Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougal
Publisher:Cengage Learning

Biochemistry
Biochemistry
ISBN:9781305577206
Author:Reginald H. Garrett, Charles M. Grisham
Publisher:Cengage Learning

Fundamentals of General, Organic, and Biological ...
Biochemistry
ISBN:9780134015187
Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. Peterson
Publisher:PEARSON