
Biochemistry
9th Edition
ISBN: 9781319114671
Author: Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.
Publisher: W. H. Freeman
expand_more
expand_more
format_list_bulleted
Question

Transcribed Image Text:### Full Amino Acid Sequence of FABP6
```
MAFTGKFEMECEKNYDEFMKLLGISSDVIEKARNFKIVTEVQQDGQDF
TWSQHYSGGHTMTNKFTVGKECNIQTMGKTFKATVQMEGGKLVVN
FPNYHQTSEIVGDKLVEVSTIGGVTYERVSKRLA
```
### Sequence Analysis
After determining the full amino acid sequence of FABP6, a BLAST search was conducted against the Protein Data Bank (PDB) to find similar structures. One structure was found to be 59% identical to the FABP6 sequence. The alignment of your protein (FABP6) is shown in blue, while the homologous structure (3ELX) is displayed in red.
**BLAST Alignment Results:**
- **Expect Score:** \(4 \times 10^{-42}\)
- **Identities:** 69/116 (50%)
- **Positives:** 88/116 (75%)
#### Sequence Alignment
```
3ELX: 5 AFNGKWETE...VTEIVQNGDDFTWTQYYPNNHVMT 64
AF GK++E EC++ Y+ F KL+GI DVI K R+FK+VTE+ Q+G D F TW+Q+Y H MT
FABP6: 2 AFTGKFMEC...TVEQQDGQDFTWSQHYSGGHTMT 61
3ELX: 65 NKF IVGKE...ISEIGGKLVE...VETSTASG 120
NKF VGKEC++T+ G FK V MEGKL+ +FP Y QT+EI G KLVE ST G
FABP6: 62 NKFTVGKE...GKT FKATVQ...VEVSTIGG 117
```
### Alignment Explanation
- **Identical Amino Acids:** Shown in black.
- **Similar, Non-identical Amino Acids:** Noted by a "+" symbol.
The sequences are aligned to show overall gaps, indicating differences in the sequences but with substantial similarities.
This alignment can help in understanding the functional and structural similarities between the two protein sequences,
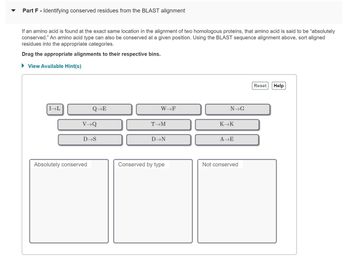
Transcribed Image Text:**Part F - Identifying conserved residues from the BLAST alignment**
If an amino acid is found at the exact same location in the alignment of two homologous proteins, that amino acid is said to be “absolutely conserved.” An amino acid type can also be conserved at a given position. Using the BLAST sequence alignment above, sort aligned residues into the appropriate categories.
**Drag the appropriate alignments to their respective bins.**
- **Amino Acid Alignments:**
- I → L
- Q → E
- W → F
- N → G
- V → Q
- T → M
- K → K
- D → S
- D → N
- A → E
**Bins:**
- Absolutely conserved
- Conserved by type
- Not conserved
**Interactive Elements:**
- Reset Button
- Help Button
_Note: This section of the webpage involves an interactive sorting activity where users classify the conservation of amino acid residues based on BLAST alignment data._
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
This is a popular solution
Trending nowThis is a popular solution!
Step by stepSolved in 4 steps with 17 images

Knowledge Booster
Similar questions
- AAAGAGAAAAGAAUA to AAAGAGAAAUGAAUA. Suppose the codon sequence has a single base pair mutation If the old protein sequence was Lys-Glu-Lys-Arg-Ile, what will be the new sequence encoded by the mutant gene? (Use the 3-letter amino acid abbreviations with hyphens and no spaces in between, i.e. Ser-Asn-Tyr-Leu-Pro.) Submit Answer Retry Entire Group No more group attempts remainarrow_forwarduestion 12 of 30 > Attempt 2 5'-GGU-3',5'-AAG-3',5'-IAC-3',5'-UGU-3', and 5'-CAA-3'. Give all A series of tRNAs have the anticodons, possible codons with which each tRNA can pair. Consider the wobble rules listed in the table. First position of anticodon C G A U I Third position of codon G U or C U A or G A, U, or C 5'-GGU-3' 5'-AAG-3' 5'-IAC-3' 5'-UGU-3' 5'-CAA-3' 3'-UUU-5' 3'-GUU-5' 3'-UUC-5' 3'-GUG-5' 3'-UUG-5' Answer Bank 3'-ACA-5' 3'-AAA-5' 3'-CUG-5' 3'-GCA-5' 3'-CCA-5' 3'-ACG-5' 3'-UGU-5' 3'-CCC-5' 3'-UCA-5' 3'-AUG-5'arrow_forwardDo you expect a Pro → Gly mutation in a surface-loop region of a globular protein to be stabilizing or destabilizing? Assume the mutant folds toa native-like conformation. Explain your answer in terms of the predictedenthalpic and entropic effects of the mutation on the ∆G for protein foldingcompared to ∆G of folding for the wild-type proteinarrow_forward
arrow_back_ios
arrow_forward_ios
Recommended textbooks for you
 BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman
BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman
Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY
Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON
Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON

Biochemistry
Biochemistry
ISBN:9781319114671
Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.
Publisher:W. H. Freeman

Lehninger Principles of Biochemistry
Biochemistry
ISBN:9781464126116
Author:David L. Nelson, Michael M. Cox
Publisher:W. H. Freeman

Fundamentals of Biochemistry: Life at the Molecul...
Biochemistry
ISBN:9781118918401
Author:Donald Voet, Judith G. Voet, Charlotte W. Pratt
Publisher:WILEY

Biochemistry
Biochemistry
ISBN:9781305961135
Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougal
Publisher:Cengage Learning

Biochemistry
Biochemistry
ISBN:9781305577206
Author:Reginald H. Garrett, Charles M. Grisham
Publisher:Cengage Learning

Fundamentals of General, Organic, and Biological ...
Biochemistry
ISBN:9780134015187
Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. Peterson
Publisher:PEARSON