
Biochemistry
9th Edition
ISBN: 9781319114671
Author: Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.
Publisher: W. H. Freeman
expand_more
expand_more
format_list_bulleted
Question
The structure of a metalloenzyme active site is down below(black picture). Describe, from a chemical and structural perspective, how the reactive site is designed to facilitate its catalytic reaction. The example below(white pitcure) suggests the level of detail that is required. Make sure that you explain what the metal is doing, what the reaction is, and its biological significance.
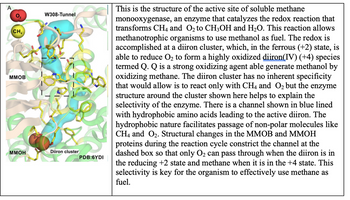
Transcribed Image Text:A
CH
MMOB
MMOH
W308-Tunnel
Diiron cluster
PDB:6YDI
This is the structure of the active site of soluble methane
monooxygenase, an enzyme that catalyzes the redox reaction that
transforms CH4 and O₂ to CH3OH and H₂O. This reaction allows
methanotrophic organisms to use methanol as fuel. The redox is
accomplished at a diiron cluster, which, in the ferrous (+2) state, is
able to reduce O₂ to form a highly oxidized diiron(IV) (+4) species
termed Q. Q is a strong oxidizing agent able generate methanol by
oxidizing methane. The diiron cluster has no inherent specificity
that would allow is to react only with CH4 and O₂ but the enzyme
structure around the cluster shown here helps to explain the
selectivity of the enzyme. There is a channel shown in blue lined
with hydrophobic amino acids leading to the active diiron. The
hydrophobic nature facilitates passage of non-polar molecules like
CH4 and O₂. Structural changes in the MMOB and MMOH
proteins during the reaction cycle constrict the channel at the
dashed box so that only O₂ can pass through when the diiron is in
the reducing +2 state and methane when it is in the +4 state. This
selectivity is key for the organism to effectively use methane as
fuel.
![A
B
172
spmoB
265
spmoBd2
spmoBd1
See this image and copyright information in PMC
Figure 1 Overall architecture of pMMO. (A) Structure of Methylocystis sp. strain M pMMO
protomer (PDB accession code 3RGR). The pmoB, pmoA, and pmoC subunits are shown in
blue, magenta, and green, respectively. The N- and C-termini of pmoB are labeled. An
exogenous helix is shown in yellow. Copper ions are shown as cyan spheres and a zinc ion is
shown as a gray sphere. Ligands are shown as ball-and-stick representations. (B) Structure of
M. capsulatus (Bath) pMMO protomer (PDB accession code 3RGB). The amino terminal
domain of pmoB (spmoBd1) is shown in blue, the carboxy terminal domain of pmoB is shown
in gray (spmoBd2), and the two transmembrane helices are shown in transparent blue. In the
recombinant spmoB protein, spmoBd1 and spmoBd2 are linked by a GKLGGG sequence
connecting residues 172 and 265 (labeled). The pmoA and pmoC subunits are shown in
transparent magenta and transparent green, respectively. A hydrophilic patch of residues
proposed to house a tricopper active site is denoted with an asterisk. The mononuclear
copper site at the interface of the two spmoB domains is not present in the Methylocystis sp.
strain M pMMO structure. [A color version of this figure is available online.]](https://content.bartleby.com/qna-images/question/d2705ec0-2034-4d28-a385-06c28bb62d2f/391b8bed-783a-4c57-aef8-6d7ec2f290bf/ye45v4b_thumbnail.png)
Transcribed Image Text:A
B
172
spmoB
265
spmoBd2
spmoBd1
See this image and copyright information in PMC
Figure 1 Overall architecture of pMMO. (A) Structure of Methylocystis sp. strain M pMMO
protomer (PDB accession code 3RGR). The pmoB, pmoA, and pmoC subunits are shown in
blue, magenta, and green, respectively. The N- and C-termini of pmoB are labeled. An
exogenous helix is shown in yellow. Copper ions are shown as cyan spheres and a zinc ion is
shown as a gray sphere. Ligands are shown as ball-and-stick representations. (B) Structure of
M. capsulatus (Bath) pMMO protomer (PDB accession code 3RGB). The amino terminal
domain of pmoB (spmoBd1) is shown in blue, the carboxy terminal domain of pmoB is shown
in gray (spmoBd2), and the two transmembrane helices are shown in transparent blue. In the
recombinant spmoB protein, spmoBd1 and spmoBd2 are linked by a GKLGGG sequence
connecting residues 172 and 265 (labeled). The pmoA and pmoC subunits are shown in
transparent magenta and transparent green, respectively. A hydrophilic patch of residues
proposed to house a tricopper active site is denoted with an asterisk. The mononuclear
copper site at the interface of the two spmoB domains is not present in the Methylocystis sp.
strain M pMMO structure. [A color version of this figure is available online.]
Expert Solution
This question has been solved!
Explore an expertly crafted, step-by-step solution for a thorough understanding of key concepts.
This is a popular solution
Trending nowThis is a popular solution!
Step by stepSolved in 2 steps with 1 images

Knowledge Booster
Similar questions
- Explain the krebs cycle. You must know the reactants, products and location for each step.arrow_forwardWhen reviewing a Michaelis-Menten Saturation Curve, at first the rate of the reaction is relatively constant, but the rate decreases as the substrate is used up and eventually reaches a plateau. Afte reaching this plateau, what would speed up the reaction again? Adding more substrate Adding heat Adding more enzyme Adding cofactorsarrow_forwardFor the three biochemical transformations below provide the names of the enzyme (or enzymes) responsible for the transformation, and identify by name or chemical structure the other necessary reactants and other products associated with each transformation. Make sure that your answer presents the missing information in a way that clearly connects it to the appropriate reaction.arrow_forward
- Which type of enzyme (Table) catalyzes the following (given) reactions?arrow_forwardEnzyme and substrate pleasearrow_forwardCatalysis through proximity and orientation effects involves: (select all that applies) Group of answer choices binding of acidic and/or basic groups in the binding site facilitating chemical reactions by binding substrates close to specific groups binding of substrate in specific, restrictive orientations depending on metal ions for catalysisarrow_forward
- A) Myth: Enzymes are specific for one substrate. Fact: Like most enzymes, alliinase can act on multiple different substrates. Explain why most enzymes can act on more than one substrate compound. (Refer to the “Alliin-like Substrates" panel in your answer.)arrow_forwardCan you please help which answers if fitting for lowering the activation energy?arrow_forwardThe structure of a metalloenzyme active site is down below(black picture). Describe, from a chemical and structural perspective, how the reactive site is designed to facilitate its catalytic reaction. The example below suggests the level of detail that is required. Make sure that you explain what the metal is doing, what the reaction is, and its biological significance.arrow_forward
- Caption: What are the biochemical properties of the following classifications of enzyme?arrow_forwardInhibitors are compounds capable of blocking the catalytic process. Outline with the use of graphs and equations in illustrating the different modes of action of enzyme inhibitors.arrow_forwardDiscuss an enzyme that acts as a catalyst in a biological system. What reaction(s) does it catalyze? What kinds of problems arise if the enzyme isn't working properly? In what ways is the enzyme's activity regulated? Other interesting facts about the enzyme? Don't forget to cite your source(s).arrow_forward
arrow_back_ios
SEE MORE QUESTIONS
arrow_forward_ios
Recommended textbooks for you
 BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman
BiochemistryBiochemistryISBN:9781319114671Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.Publisher:W. H. Freeman Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman
Lehninger Principles of BiochemistryBiochemistryISBN:9781464126116Author:David L. Nelson, Michael M. CoxPublisher:W. H. Freeman Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY
Fundamentals of Biochemistry: Life at the Molecul...BiochemistryISBN:9781118918401Author:Donald Voet, Judith G. Voet, Charlotte W. PrattPublisher:WILEY BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305961135Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougalPublisher:Cengage Learning BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning
BiochemistryBiochemistryISBN:9781305577206Author:Reginald H. Garrett, Charles M. GrishamPublisher:Cengage Learning Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON
Fundamentals of General, Organic, and Biological ...BiochemistryISBN:9780134015187Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. PetersonPublisher:PEARSON

Biochemistry
Biochemistry
ISBN:9781319114671
Author:Lubert Stryer, Jeremy M. Berg, John L. Tymoczko, Gregory J. Gatto Jr.
Publisher:W. H. Freeman

Lehninger Principles of Biochemistry
Biochemistry
ISBN:9781464126116
Author:David L. Nelson, Michael M. Cox
Publisher:W. H. Freeman

Fundamentals of Biochemistry: Life at the Molecul...
Biochemistry
ISBN:9781118918401
Author:Donald Voet, Judith G. Voet, Charlotte W. Pratt
Publisher:WILEY

Biochemistry
Biochemistry
ISBN:9781305961135
Author:Mary K. Campbell, Shawn O. Farrell, Owen M. McDougal
Publisher:Cengage Learning

Biochemistry
Biochemistry
ISBN:9781305577206
Author:Reginald H. Garrett, Charles M. Grisham
Publisher:Cengage Learning

Fundamentals of General, Organic, and Biological ...
Biochemistry
ISBN:9780134015187
Author:John E. McMurry, David S. Ballantine, Carl A. Hoeger, Virginia E. Peterson
Publisher:PEARSON